2PIO
 
 | | Androgen receptor LBD with small molecule | | Descriptor: | 2-METHYL-1H-INDOLE, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Estebanez-Perpina, E, Arnold, A.A, Baxter, J.D, Webb, P, Guy, R.K, Fletterick, K.R. | | Deposit date: | 2007-04-13 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A surface on the androgen receptor that allosterically regulates coactivator binding.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PIV
 
 | | Androgen receptor with small molecule | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, ... | | Authors: | Estebanez-Perpina, E, Arnold, A.A, Baxter, J.D, Webb, P, Guy, R.K, Fletterick, K.R. | | Deposit date: | 2007-04-13 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A surface on the androgen receptor that allosterically regulates coactivator binding.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PIU
 
 | | Androgen receptor LBD with small molecule | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, [4-(4-HYDROXY-3-IODO-PHENOXY)-3,5-DIIODO-PHENYL]-ACETIC ACID | | Authors: | Estebanez-Perpina, E, Arnold, A.A, Baxter, J.D, Webb, P, Guy, R.K, Fletterick, K.R. | | Deposit date: | 2007-04-13 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A surface on the androgen receptor that allosterically regulates coactivator binding.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PIT
 
 | | Androgen receptor LBD with small molecule | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, SULFATE ION, ... | | Authors: | Estebanez-Perpina, E, Arnold, A.A, Baxter, J.D, Webb, P, Guy, R.K, Fletterick, K.R. | | Deposit date: | 2007-04-13 | | Release date: | 2007-09-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A surface on the androgen receptor that allosterically regulates coactivator binding.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PIP
 
 | | Androgen receptor LBD with small molecule | | Descriptor: | 1-TERT-BUTYL-3-(2,5-DIMETHYLBENZYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-AMINE, 1H-INDOLE-3-CARBOXYLIC ACID, 5-ALPHA-DIHYDROTESTOSTERONE, ... | | Authors: | Estebanez-Perpina, E, Arnold, A.A, Baxter, J.D, Webb, P, Guy, R.K, Fletterick, K.R. | | Deposit date: | 2007-04-13 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A surface on the androgen receptor that allosterically regulates coactivator binding.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2QPY
 
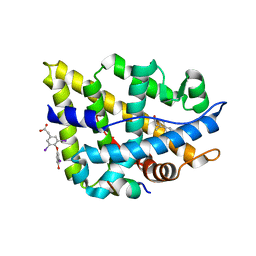 | | AR LBD with small molecule | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Coactivator peptide, ... | | Authors: | Estebanez-Perpina, E, Fletterick, R. | | Deposit date: | 2007-07-25 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A surface on the androgen receptor that allosterically regulates coactivator binding.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PIX
 
 | | AR LBD with small molecule | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor | | Authors: | Estebanez-Perpina, E, Arnold, A.A, Baxter, J.D, Webb, P, Guy, R.K, Fletterick, K.R. | | Deposit date: | 2007-04-13 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A surface on the androgen receptor that allosterically regulates coactivator binding.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2HH0
 
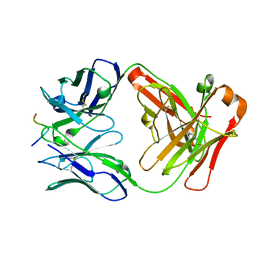 | |
4FRZ
 
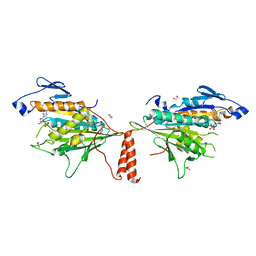 | |
1DPO
 
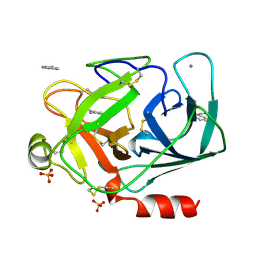 | | STRUCTURE OF RAT TRYPSIN | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Stroud, R.M. | | Deposit date: | 1997-03-31 | | Release date: | 1997-07-07 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | 1.59 A structure of trypsin at 120 K: comparison of low temperature and room temperature structures.
Proteins, 10, 1991
|
|
4QK4
 
 | | Crystal structure of human nuclear receptor sf-1 (nr5a1) bound to pip2 at 2.8 a resolution | | Descriptor: | (2S)-3-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dihexadecanoate, 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2014-06-05 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4RBO
 
 | |
4QJR
 
 | | Crystal structure of human nuclear receptor sf-1 (nr5a1) bound to its hormone pip3 at 2.4 a resolution | | Descriptor: | (2S)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dihexadecanoate, ACETATE ION, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Joint Center for Structural Genomics (JCSG), Partnership for Stem Cell Biology (STEMCELL) | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3U21
 
 | |
1ME3
 
 | |
1ME4
 
 | |
8GPB
 
 | |
5GPB
 
 | |
3H4S
 
 | |
2GPB
 
 | |
4GPB
 
 | |
1YOW
 
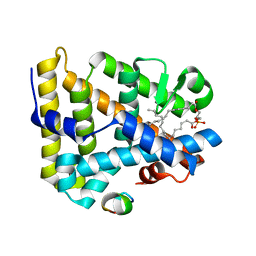 | | human Steroidogenic Factor 1 LBD with bound Co-factor Peptide | | Descriptor: | PHOSPHATIDYL ETHANOL, Steroidogenic factor 1, TIF2 peptide | | Authors: | Krylova, I.N, Sablin, E.P, Xu, R.X, Waitt, G.M, Juzumiene, D, Williams, J.D, Ingraham, H.A, Willson, T.M, Williams, S.P, Montana, V, Madauss, K.P, Moore, J, Bynum, J.M, Lebedeva, L, MacKay, J.A, Suzawa, M, Guy, R.K, Thornton, J.W. | | Deposit date: | 2005-01-28 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1
Cell(Cambridge,Mass.), 120, 2005
|
|
1YVB
 
 | | the Plasmodium falciparum Cysteine Protease Falcipain-2 | | Descriptor: | Cystatin, GLYCEROL, falcipain 2 | | Authors: | Wang, S.X. | | Deposit date: | 2005-02-15 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for unique mechanisms of folding and hemoglobin binding by a malarial protease.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3GPB
 
 | |
1ST3
 
 | |
