6FPK
 
 | | Co-translational folding intermediate dictates membrane targeting of the signal recognition particle (SRP)- receptor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Signal recognition particle receptor FtsY | | Authors: | Karniel, A, Mrusek, D, Steinchen, W, Dym, O, Bange, G, Bibi, E. | | Deposit date: | 2018-02-11 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Co-translational Folding Intermediate Dictates Membrane Targeting of the Signal Recognition Particle Receptor.
J. Mol. Biol., 430, 2018
|
|
6FPR
 
 | | Co-translational folding intermediate dictates membrane targeting of the signal recognition particle (SRP)- receptor | | Descriptor: | Signal recognition particle receptor FtsY | | Authors: | Karniel, A, Mrusek, D, Steinchen, W, Dym, O, Bange, G, Bibi, E. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Co-translational Folding Intermediate Dictates Membrane Targeting of the Signal Recognition Particle Receptor.
J. Mol. Biol., 430, 2018
|
|
6FPF
 
 | |
6G12
 
 | | Crystal structure of GMPPNP bound RbgA from S. aureus | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ribosome biogenesis GTPase A | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA.
J. Biol. Chem., 293, 2018
|
|
6G14
 
 | | Crystal structure of ppGpp bound RbgA from S. aureus | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, Ribosome biogenesis GTPase A | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA.
J. Biol. Chem., 293, 2018
|
|
6G0Z
 
 | | Crystal structure of GDP bound RbgA from S. aureus | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ribosome biogenesis GTPase A | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA.
J. Biol. Chem., 293, 2018
|
|
6FPG
 
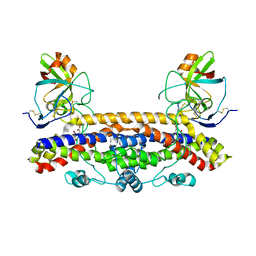 | | Structure of the Ustilago maydis chorismate mutase 1 in complex with a Zea mays kiwellin | | Descriptor: | CITRIC ACID, Chromosome 16, whole genome shotgun sequence, ... | | Authors: | Altegoer, F, Steinchen, W, Bange, G. | | Deposit date: | 2018-02-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A kiwellin disarms the metabolic activity of a secreted fungal virulence factor.
Nature, 565, 2019
|
|
6G15
 
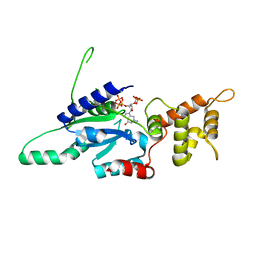 | | Crystal structure of pppGpp bound RbgA from S. aureus | | Descriptor: | Ribosome biogenesis GTPase A, guanosine 5'-(tetrahydrogen triphosphate) 3'-(trihydrogen diphosphate) | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-03-20 | | Release date: | 2018-11-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA.
J. Biol. Chem., 293, 2018
|
|
6HJW
 
 | |
6H3P
 
 | |
6H9H
 
 | | Csf5, CRISPR-Cas type IV Cas6 crRNA endonuclease | | Descriptor: | Csf5, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-08-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum.
Nat Microbiol, 4, 2019
|
|
6H9I
 
 | | Csf5, CRISPR-Cas type IV Cas6 crRNA endonuclease | | Descriptor: | Csf5, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Pausch, P, Bange, G. | | Deposit date: | 2018-08-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum.
Nat Microbiol, 4, 2019
|
|
6GOW
 
 | |
8BEI
 
 | | Structure of hexameric subcomplexes (Truncation Delta2-6) of the fractal citrate synthase from Synechococcus elongatus PCC7942 | | Descriptor: | Citrate synthase | | Authors: | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | Deposit date: | 2022-10-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Emergence of fractal geometries in the evolution of a metabolic enzyme.
Nature, 628, 2024
|
|
7A6S
 
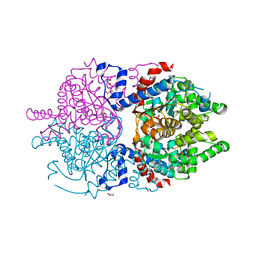 | | Crystal Structure of Asn173Ser variant of Human Deoxyhypusine Synthase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Deoxyhypusine synthase, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-08-26 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cryo-EM structure of human eIF5A-DHS complex reveals the molecular basis of hypusination-associated neurodegenerative disorders.
Nat Commun, 14, 2023
|
|
7A6T
 
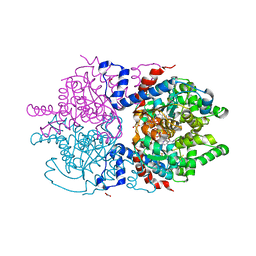 | | Crystal Structure of Asn173Ser variant of Human Deoxyhypusine Synthase in complex with NAD and spermidine | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Deoxyhypusine synthase, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-08-26 | | Release date: | 2022-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Cryo-EM structure of human eIF5A-DHS complex reveals the molecular basis of hypusination-associated neurodegenerative disorders.
Nat Commun, 14, 2023
|
|
7AFT
 
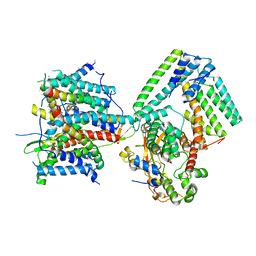 | | Cryo-EM structure of the signal sequence-engaged post-translational Sec translocon | | Descriptor: | Mating factor alpha-1,Mating factor alpha-1, Protein translocation protein SEC63, Protein transport protein SBH1, ... | | Authors: | Weng, T.-H, Beatrix, B, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-09-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Architecture of the active post-translational Sec translocon.
Embo J., 40, 2021
|
|
3IQX
 
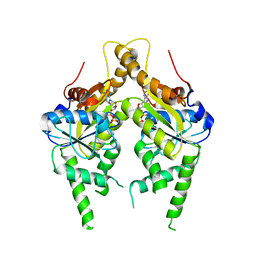 | | ADP complex of C.therm. Get3 in closed form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Tail-anchored protein targeting factor Get3, ... | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2009-08-21 | | Release date: | 2009-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insights into tail-anchored protein binding and membrane insertion by Get3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IQW
 
 | | AMPPNP complex of C. therm. Get3 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tail-anchored protein targeting factor Get3, ... | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2009-08-21 | | Release date: | 2009-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into tail-anchored protein binding and membrane insertion by Get3.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4NWB
 
 | | Crystal structure of Mrt4 | | Descriptor: | SULFATE ION, mRNA turnover protein 4 | | Authors: | Holdermann, I, Sinning, I. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
6SBW
 
 | | CdbA Form One | | Descriptor: | CdbA | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2019-07-22 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | CdbA is a DNA-binding protein and c-di-GMP receptor important for nucleoid organization and segregation in Myxococcus xanthus.
Nat Commun, 11, 2020
|
|
4ZN4
 
 | |
6SBX
 
 | | CdbA Form Two | | Descriptor: | CdbA | | Authors: | Lovering, A.L, Cadby, I.T. | | Deposit date: | 2019-07-22 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | CdbA is a DNA-binding protein and c-di-GMP receptor important for nucleoid organization and segregation in Myxococcus xanthus.
Nat Commun, 11, 2020
|
|
8QZP
 
 | | Structure of the non-mitochondrial citrate synthase from Ananas comosus | | Descriptor: | Citrate synthase | | Authors: | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | Deposit date: | 2023-10-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Frequent transitions in self-assembly across the evolution of a central metabolic enzyme.
Biorxiv, 2024
|
|
8RJL
 
 | | Structure of a first order Sierpinski triangle formed by the H369R mutant of the citrate synthase from Synechococcus elongatus | | Descriptor: | Citrate synthase | | Authors: | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Emergence of fractal geometries in the evolution of a metabolic enzyme.
Nature, 628, 2024
|
|
