6L7O
 
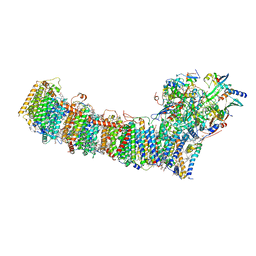 | | cryo-EM structure of cyanobacteria Fd-NDH-1L complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Zhang, C, Shuai, J, Wu, J, Lei, M. | | Deposit date: | 2019-11-02 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into NDH-1 mediated cyclic electron transfer.
Nat Commun, 11, 2020
|
|
6LBS
 
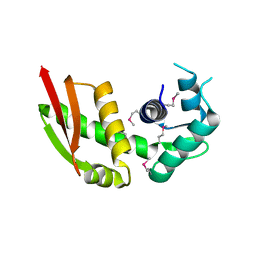 | | Crystal structure of yeast Stn1 | | Descriptor: | KLLA0C11825p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LBR
 
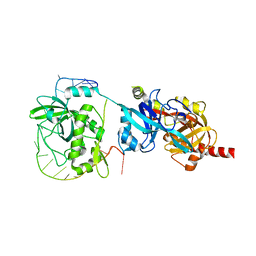 | | Crystal structure of yeast Cdc13 and ssDNA | | Descriptor: | KLLA0F20922p, Telomere single-strand DNA | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LBT
 
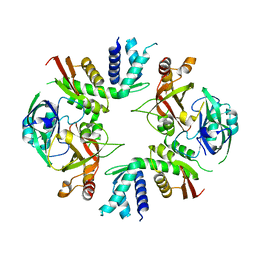 | | Crystal structure of yeast Cdc13 and Stn1 | | Descriptor: | GLYCEROL, KLLA0C11825p, KLLA0F20922p,KLLA0F20922p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LBU
 
 | | Crystal structure of yeast Stn1 and Ten1 | | Descriptor: | KLLA0C11825p, KLLA0E09417p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6L7P
 
 | | cryo-EM structure of cyanobacteria NDH-1LdelV complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, ... | | Authors: | Zhang, C, Shuai, J, Wu, J, Lei, M. | | Deposit date: | 2019-11-02 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into NDH-1 mediated cyclic electron transfer.
Nat Commun, 11, 2020
|
|
6MLC
 
 | | PHD6 domain of MLL3 in complex with histone H4 | | Descriptor: | GLYCEROL, Histone H4, Histone-lysine N-methyltransferase 2C, ... | | Authors: | Dong, A, Liu, Y, Qin, S, Lei, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into trans-histone regulation of H3K4 methylation by unique histone H4 binding of MLL3/4.
Nat Commun, 10, 2019
|
|
7C7A
 
 | | Cryo-EM structure of yeast Ribonuclease MRP with substrate ITS1 | | Descriptor: | Internal Transcribed Spacer 1, MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, ... | | Authors: | Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-05-24 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insight into precursor ribosomal RNA processing by ribonuclease MRP.
Science, 369, 2020
|
|
7C79
 
 | | Cryo-EM structure of yeast Ribonuclease MRP | | Descriptor: | MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, Ribonuclease MRP RNA subunit NME1, ... | | Authors: | Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-05-24 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insight into precursor ribosomal RNA processing by ribonuclease MRP.
Science, 369, 2020
|
|
7D59
 
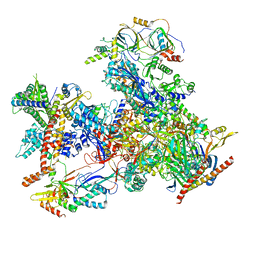 | | cryo-EM structure of human RNA polymerase III in apo state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D58
 
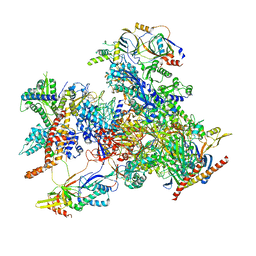 | | cryo-EM structure of human RNA polymerase III in elongating state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7WTD
 
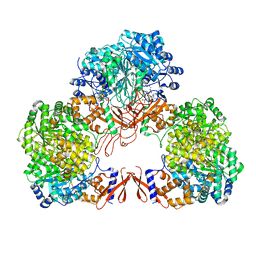 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA in the intermediate state 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, COENZYME A, Pyruvate carboxylase, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTE
 
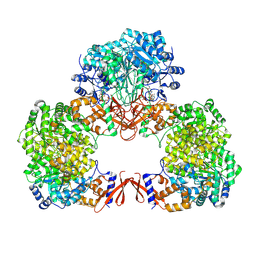 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA in the intermediate state 2 | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-TRIPHOSPHATE, Pyruvate carboxylase, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTA
 
 | | Cryo-EM structure of human pyruvate carboxylase in apo state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Pyruvate carboxylase, mitochondrial | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTC
 
 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA in the ground state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ACETYL COENZYME *A, Pyruvate carboxylase, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7WTB
 
 | | Cryo-EM structure of human pyruvate carboxylase with acetyl-CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ACETYL COENZYME *A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chai, P, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2022-02-04 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanistic insight into allosteric activation of human pyruvate carboxylase by acetyl-CoA.
Mol.Cell, 82, 2022
|
|
7X5C
 
 | | Solution structure of Tetrahymena p75OB1-p50PBM | | Descriptor: | Telomerase associated protein p50PBM, Telomerase-associated protein p75OB1 | | Authors: | Wu, B, Tang, T, Xue, H.J, Wu, J, Lei, M. | | Deposit date: | 2022-03-04 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Association of the CST complex and p50 in Tetrahymena is crucial for telomere maintenance
Structure, 2022
|
|
7E34
 
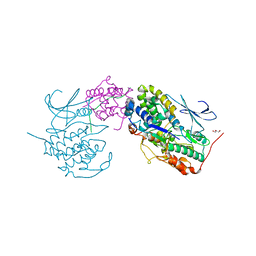 | | Crystal structure of SUN1-Speedy A-CDK2 | | Descriptor: | Cyclin-dependent kinase 2, GLYCEROL, SUN domain-containing protein 1, ... | | Authors: | Chen, Y, Huang, C, Wu, J, Lei, M. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The SUN1-SPDYA interaction plays an essential role in meiosis prophase I.
Nat Commun, 12, 2021
|
|
7D60
 
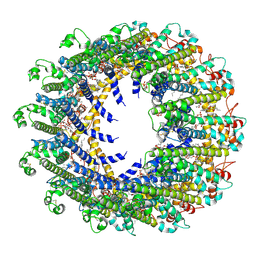 | | Cryo-EM Structure of human CALHM5 in the presence of rubidium red | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | Authors: | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
7CUJ
 
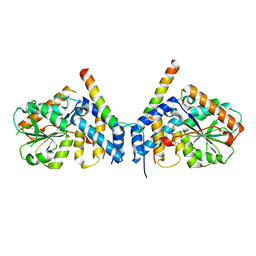 | | Crystal structure of fission yeast Ccq1 and Tpz1 | | Descriptor: | Coiled-coil quantitatively-enriched protein 1, Protection of telomeres protein tpz1 | | Authors: | Sun, H, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2020-08-23 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
7CUH
 
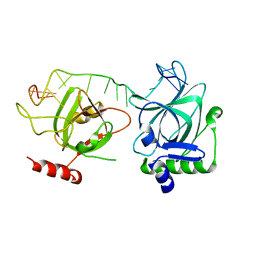 | | Crystal structure of fission yeast Pot1 and ssDNA | | Descriptor: | Protection of telomeres protein 1, Telomere single-strand DNA | | Authors: | Sun, H, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2020-08-23 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
7CUI
 
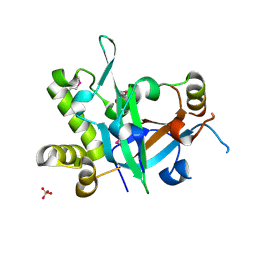 | | Crystal structure of fission yeast Pot1 and Tpz1 | | Descriptor: | Protection of telomeres protein 1, Protection of telomeres protein tpz1, SULFATE ION | | Authors: | Sun, H, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2020-08-23 | | Release date: | 2021-08-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into Pot1-ssDNA, Pot1-Tpz1 and Tpz1-Ccq1 Interactions within fission yeast shelterin complex.
Plos Genet., 18, 2022
|
|
7D65
 
 | | Cryo-EM Structure of human CALHM5 in the presence of Ca2+ | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | Authors: | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
7D61
 
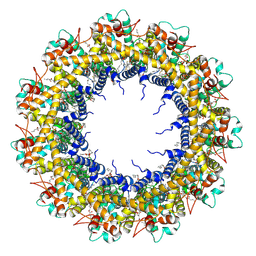 | | Cryo-EM Structure of human CALHM5 in the presence of EDTA | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, Calcium homeostasis modulator protein 5 | | Authors: | Liu, J, Guan, F.H, Wu, J, Wan, F.T, Lei, M, Ye, S. | | Deposit date: | 2020-09-28 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of human calcium homeostasis modulator 5.
Cell Discov, 6, 2020
|
|
7X9G
 
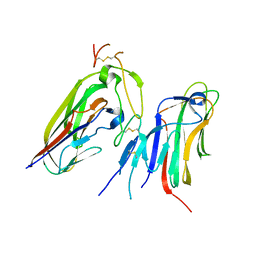 | | Crystal structure of human EDA and EDAR | | Descriptor: | Ectodysplasin-A, secreted form, Tumor necrosis factor receptor superfamily member EDAR | | Authors: | Yu, K, Wan, F, Huang, C, Wu, J, Lei, M. | | Deposit date: | 2022-03-15 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into pathogenic mechanism of hypohidrotic ectodermal dysplasia caused by ectodysplasin A variants.
Nat Commun, 14, 2023
|
|
