4CY6
 
 | | apo structure of 2-hydroxybiphenyl 3-monooxygenase HbpA | | Descriptor: | 2-HYDROXYBIPHENYL-3-MONOOXYGENASE | | Authors: | Jensen, C.N, Farrugia, J.E, Frank, A, Man, H, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2014-04-10 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structures of the Apo and Fad-Bound Forms of 2-Hydroxybiphenyl 3-Monooxygenase (Hbpa) Locate Activity Hotspots Identified by Using Directed Evolution.
Chembiochem, 16, 2015
|
|
4D3S
 
 | | Imine reductase from Nocardiopsis halophila | | Descriptor: | IMINE REDUCTASE, octyl beta-D-glucopyranoside | | Authors: | Man, H, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2014-10-23 | | Release date: | 2015-04-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure, Activity and Stereoselectivity of Nadph-Dependent Oxidoreductases Catalysing the S-Selective Reduction of the Imine Substrate 2-Methylpyrroline.
Chembiochem, 16, 2015
|
|
4BMV
 
 | | Short-chain dehydrogenase from Sphingobium yanoikuyae in complex with NADPH | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SHORT-CHAIN DEHYDROGENASE | | Authors: | Man, H, Kedziora, K, Lavandera-Garcia, I, Gotor-Fernandez, V, Grogan, G. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Alcohol Dehydrogenases from Ralstonia and Sphingobium Spp. Reveal the Molecular Basis for Their Recognition of 'Bulky-Bulky' Ketones
Top.Catal., 57, 2014
|
|
4BMN
 
 | | apo structure of short-chain alcohol dehydrogenase from Ralstonia sp. DSM 6428 | | Descriptor: | 1,2-ETHANEDIOL, ALCLOHOL DEHYDROGENASE/SHORT-CHAIN DEHYDROGENASE, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Man, H, Kulig, J, Rother, D, Grogan, G. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of Alcohol Dehydrogenases from Ralstonia and Sphingobium Spp. Reveal the Molecular Basis for Their Recognition of 'Bulky-Bulky' Ketones
Top.Catal., 57, 2014
|
|
4C4O
 
 | | Structure of carbonyl reductase CPCR2 from Candida parapsilosis in complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, CARBONYL REDUCTASE CPCR2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Man, H, Loderer, C, Ansorge-Schumacher, M, Grogan, G. | | Deposit date: | 2013-09-06 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Nadh-Dependent Carbonyl Reductase (Cpcr2) from Candida Parapsilosis Provides Insight Into Mutations that Improve Catalytic Properties
Chemcatchem, 6, 2014
|
|
4C5O
 
 | | Flavin monooxygenase from Stenotrophomonas maltophilia. Q193R H194T mutant | | Descriptor: | FLAVIN MONOOXYGENASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Jensen, C.N, Ali, S.T, Allen, M.J, Grogan, G. | | Deposit date: | 2013-09-13 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutations of an Nad(P)H-Dependent Flavoprotein Monooxygenase that Influence Cofactor Promiscuity and Enantioselectivity.
FEBS Open Bio, 3, 2013
|
|
4BMS
 
 | | Short chain alcohol dehydrogenase from Ralstonia sp. DSM 6428 in complex with NADPH | | Descriptor: | ALCLOHOL DEHYDROGENASE/SHORT-CHAIN DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Man, H, Kulig, J, Rother, D, Grogan, G. | | Deposit date: | 2013-05-10 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structures of Alcohol Dehydrogenases from Ralstonia and Sphingobium Spp. Reveal the Molecular Basis for Their Recognition of 'Bulky-Bulky' Ketones
Top.Catal., 57, 2014
|
|
5FWN
 
 | | Imine Reductase from Amycolatopsis orientalis. Closed form in in complex with (R)- Methyltetrahydroisoquinoline | | Descriptor: | (1R)-1-methyl-1,2,3,4-tetrahydroisoquinoline, IMINE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | Deposit date: | 2016-02-18 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Stereoselectivity and Structural Characterization of an Imine Reductase (Ired) from Amycolatopsis Orientalis
Acs Catalysis, 6, 2016
|
|
5G6R
 
 | | Imine reductase from Aspergillus oryzae | | Descriptor: | IMINE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | Deposit date: | 2016-06-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A reductive aminase from Aspergillus oryzae.
Nat Chem, 9, 2017
|
|
5G6S
 
 | | Imine reductase from Aspergillus oryzae in complex with NADP(H) and (R)-rasagiline | | Descriptor: | IMINE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RASAGILINE | | Authors: | Man, H, Aleku, G, Turner, N.J, Grogan, G. | | Deposit date: | 2016-06-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A reductive aminase from Aspergillus oryzae.
Nat Chem, 9, 2017
|
|
5FYF
 
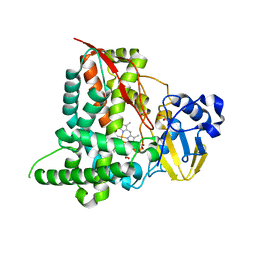 | | Structure of CYP153A from Marinobacter aquaeolei | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Danesh Azari, H.R, Spandolf, C, Hoffman, S.M, Weissenborn, M, Hauer, B, Grogan, G. | | Deposit date: | 2016-03-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-Guided Redesign of CYP153AM.aqfor the Improved Terminal Hydroxylation of Fatty Acids
Chemcatchem, 2016
|
|
5FR9
 
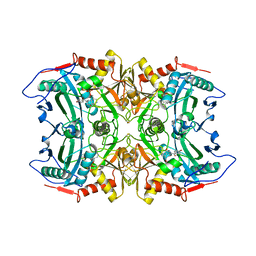 | | Structure of transaminase ATA-117 arRmut11 from Arthrobacter sp. KNK168 inhibited with 1-(4-Bromophenyl)-2-fluoroethylamine | | Descriptor: | (R)-AMINE TRANSAMINASE, [4-[3-(4-bromophenyl)-3-oxidanylidene-propyl]-6-methyl-5-oxidanyl-pyridin-3-yl]methyl phosphate | | Authors: | Cuetos, A, Kroutil, W, Lavandera, I, Grogan, G. | | Deposit date: | 2015-12-16 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Catalytic Promiscuity of Transaminases: Preparation of Enantioenriched Beta-Fluoroamines by Formal Tandem Hydrodefluorination/Deamination.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5FYG
 
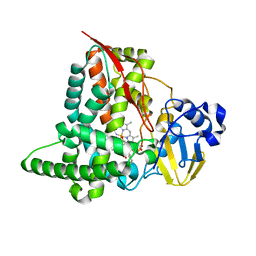 | | Structure of CYP153A from Marinobacter aquaeolei in complex with hydroxydodecanoic acid | | Descriptor: | 12-HYDROXYDODECANOIC ACID, CYTOCHROME P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Danesh-Azari, H.-R, Spandolf, C, Hoffman, S.M, Weissenborn, M, Hauer, B, Grogan, G. | | Deposit date: | 2016-03-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-Guided Redesign of CYP153AM.aqfor the Improved Terminal Hydroxylation of Fatty Acids
Chemcatchem, 2016
|
|
5G1W
 
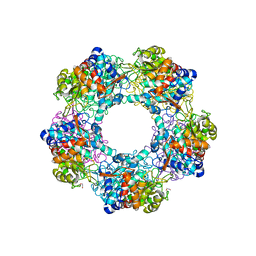 | | Apo Structure of Linalool Dehydratase-Isomerase | | Descriptor: | 1,2-ETHANEDIOL, LINALOOL DEHYDRATASE/ISOMERASE, METHYLMALONIC ACID | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
5G1U
 
 | | Linalool Dehydratase Isomerase in complex with Geraniol | | Descriptor: | Geraniol, LINALOOL DEHYDRATASE/ISOMERASE | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
5G1V
 
 | | Linalool Dehydratase Isomerase: Selenomethionine Derivative | | Descriptor: | LINALOOL DEHYDRATASE ISOMERASE | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
5G4I
 
 | | PLP-dependent phospholyase A1RDF1 from Arthrobacter aurescens TC1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Cuetos, A, Tuan, A.N, Mangas Sanchez, J, Grogan, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-19 | | Last modified: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Phospholyase Activity of a Class III Transaminase Homologue.
Chembiochem, 17, 2016
|
|
5G4J
 
 | | Phospholyase A1RDF1 from Arthrobacter in complex with phosphoethanolamine | | Descriptor: | PUTATIVE AMINOTRANSFERASE CLASS III PROTEIN, SODIUM ION, {5-hydroxy-6-methyl-4-[(E)-{[2-(phosphonooxy)ethyl]imino}methyl]pyridin-3-yl}methyl dihydrogen phosphate | | Authors: | Cuetos, A, Tuan, A.N, Mangas Sanchez, J, Grogan, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for Phospholyase Activity of a Class III Transaminase Homologue.
Chembiochem, 17, 2016
|
|
5G5A
 
 | |
7OSN
 
 | | IRED361 from Micromonospora sp. in complex with NADP+ | | Descriptor: | 6-phosphogluconate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gilio, A.K, Harawa, V, Turner, N, Grogan, G.J. | | Deposit date: | 2021-06-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Synthesis of Stereoenriched Piperidines via Chemo-Enzymatic Dearomatization of Activated Pyridines.
J.Am.Chem.Soc., 144, 2022
|
|
8BK1
 
 | | Mutant Imine Reductase IR007-143 from Amycolatopsis azurea, E120A, M197W, M206S, A213P, D238G, I240L | | Descriptor: | Mutant Imine Reductase IR007-143 from Amycolatopsis azurea, E120A, M197W, ... | | Authors: | Gilio, A.K, Grogan, G.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineering of a Reductive Aminase to Enable the Synthesis of a Key Intermediate to a CDK 2/4/6 Inhibitor
Acs Catalysis, 13, 2023
|
|
3JV7
 
 | | Structure of ADH-A from Rhodococcus ruber | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ADH-A, ... | | Authors: | Karabec, M, Lyskowski, A, Gruber, K. | | Deposit date: | 2009-09-16 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into substrate specificity and solvent tolerance in alcohol dehydrogenase ADH-'A' from Rhodococcus ruber DSM 44541.
Chem.Commun.(Camb.), 2010
|
|
2WQ8
 
 | | Glycan labelling using engineered variants of galactose oxidase obtained by directed evolution | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Rannes, J.G, Ioannou, A, Willies, S.C, Behrens, C, Grogan, G.J, Flitsch, S.L, Turner, N.J. | | Deposit date: | 2009-08-14 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Glycoprotein Labeling Using Engineered Variants of Galactose Oxidase Obtained by Directed Evolution.
J.Am.Chem.Soc., 133, 2011
|
|
