4EEN
 
 | | crystal structure of HAD FAMILY HYDROLASE DR_1622 from Deinococcus radiodurans R1 (TARGET EFI-501256) with bound magnesium | | Descriptor: | Beta-phosphoglucomutase-related protein, CHLORIDE ION, MAGNESIUM ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-28 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of HAD HYDROLASE DR_1622 Deinococcus radiodurans R1 (TARGET EFI-501256)
To be Published
|
|
3GMG
 
 | | Crystal structure of an uncharacterized conserved protein from Mycobacterium tuberculosis | | Descriptor: | Uncharacterized protein Rv1825/MT1873 | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized conserved protein from Mycobacterium tuberculosis
To be Published
|
|
3GMS
 
 | |
4GLO
 
 | | Crystal structure of a short chain dehydrogenase homolog (target EFI-505321) from burkholderia multivorans, with bound NAD | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier protein] reductase, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Hobbs, M.E, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-14 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a short chain dehydrogenase homolog
(target EFI-505321) from burkholderia multivorans, with bound NAD
To be Published
|
|
3GY0
 
 | | Crystal structure of putatitve short chain dehydrogenase FROM SHIGELLA FLEXNERI 2A STR. 301 complexed with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of putatitve short chain dehydrogenase
FROM SHIGELLA FLEXNERI 2A STR. 301 complexed with NADP
To be Published
|
|
4GLT
 
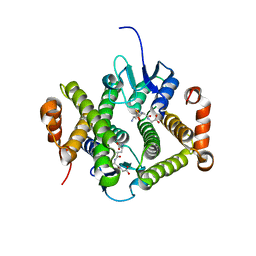 | | Crystal structure of glutathione s-transferase MFLA_2116 (target EFI-507160) from methylobacillus flagellatus kt with gsh bound | | Descriptor: | GLUTATHIONE, Glutathione S-transferase-like protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-14 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione s-transferase MFLA_2116 (target EFI-507160) from methylobacillus flagellatus kt with gsh bound
To be Published
|
|
4GEK
 
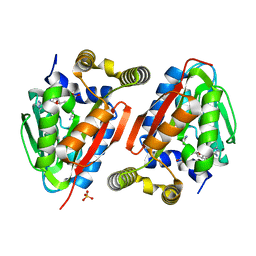 | | Crystal Structure of wild-type CmoA from E.coli | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, SULFATE ION, tRNA (cmo5U34)-methyltransferase | | Authors: | Kim, J, Toro, R, Bonanno, J.B, Bhosle, R, Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-02 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function
Nature, 498, 2013
|
|
3GKB
 
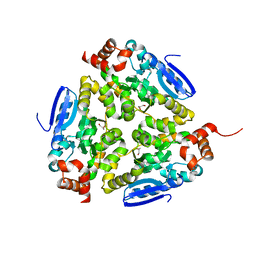 | | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis | | Descriptor: | GLYCEROL, Putative enoyl-CoA hydratase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-10 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase from Streptomyces avermitilis
To be Published
|
|
3GNL
 
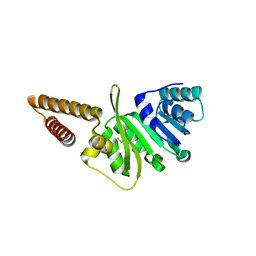 | | Structure of uncharacterized protein (LMOf2365_1472) from Listeria monocytogenes serotype 4b | | Descriptor: | uncharacterized protein, DUF633, LMOf2365_1472 | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-17 | | Release date: | 2009-04-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of uncharacterized protein (LMOf2365_1472) from
Listeria monocytogenes serotype 4b
To be Published
|
|
3GP4
 
 | | Crystal structure of putative MerR family transcriptional regulator from listeria monocytogenes | | Descriptor: | D-METHIONINE, GLYCEROL, SODIUM ION, ... | | Authors: | Ramagopal, U.A, Toro, R, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-20 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of putative MerR family transcriptional regulator from Listeria monocytogenes
To be published
|
|
3GT7
 
 | | CRYSTAL STRUCTURE OF SIGNAL RECEIVER DOMAIN OF SIGNAL TRANSDUCTION HISTIDINE KINASE FROM Syntrophus aciditrophicus | | Descriptor: | Sensor protein | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Freeman, J, Hu, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Signal Transduction Kinase from Syntrophus Aciditrophicus
To be Published
|
|
4FB5
 
 | |
4FHQ
 
 | | Crystal Structure of HVEM | | Descriptor: | Tumor necrosis factor receptor superfamily member 14 | | Authors: | Liu, W, Zhan, C, Patskovsky, Y, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Increased Heterologous Protein Expression in Drosophila S2 Cells for Massive Production of Immune Ligands/Receptors and Structural Analysis of Human HVEM.
Mol Biotechnol, 57, 2015
|
|
4DIO
 
 | | The crystal structure of transhydrogenase from Sinorhizobium meliloti | | Descriptor: | NAD(P) transhydrogenase subunit alpha part 1 | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-01-31 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of transhydrogenase from Sinorhizobium meliloti
To be Published
|
|
4DND
 
 | | Crystal structure of syntaxin 10 from Homo sapiens | | Descriptor: | Syntaxin-10 | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-08 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of syntaxin 10 from Homo sapiens
To be Published
|
|
4DIA
 
 | | CRYSTAL STRUCTURE OF THE D248N mutant of 2-PYRONE-4,6-DICARBOXYLIC ACID HYDROLASE FROM SPHINGOMONAS PAUCIMOBILIS complexed with substrate at pH 4.6 | | Descriptor: | 2-pyrone-4,6-dicarbaxylate hydrolase | | Authors: | Malashkevich, V.N, Toro, R, Hobbs, M.E, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-01-11 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
4DA9
 
 | | Crystal structure of putative Short-chain dehydrogenase/reductase from Sinorhizobium meliloti 1021 | | Descriptor: | SULFATE ION, Short-chain dehydrogenase/reductase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-01-12 | | Release date: | 2012-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative Short-chain dehydrogenase/reductase from Sinorhizobium meliloti 1021
To be Published
|
|
4DNA
 
 | | CRYSTAL STRUCTURE OF putative glutathione reductase from Sinorhizobium meliloti 1021 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Probable glutathione reductase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-08 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE OF putative glutathione reductase from Sinorhizobium meliloti 1021
To be Published
|
|
4DRY
 
 | | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase from Rhizobium meliloti | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, SULFATE ION | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-02-17 | | Release date: | 2012-02-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase from Rhizobium meliloti
To be Published
|
|
3IVR
 
 | | CRYSTAL STRUCTURE OF PUTATIVE long-chain-fatty-acid CoA ligase FROM Rhodopseudomonas palustris CGA009 | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative long-chain-fatty-acid CoA ligase | | Authors: | Patskovsky, Y, Toro, R, Foti, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-01 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF PUTATIVE long-chain-fatty-acid CoA SYNTHASE FROM Rhodopseudomonas palustris CGA009
To be Published
|
|
4DLM
 
 | | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P212121 | | Descriptor: | Amidohydrolase 2, ZINC ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Seidel, R.D, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Al Obaidi, N.F, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Crystal structure of an amidohydrolase (COG3618) from burkholderia multivorans (TARGET EFI-500235) with bound ZN, space group P212121
to be published
|
|
3IVE
 
 | | Putative 5'-Nucleotidase (c4898) from Escherichia Coli in complex with Cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CHLORIDE ION, FE (III) ION, ... | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Putative 5'-Nucleotidase (c4898) from Escherichia Coli in complex with Cytidine
To be Published
|
|
4EUM
 
 | | Crystal structure of a sugar kinase (Target EFI-502132) from Oceanicola granulosus with bound AMP, crystal form II | | Descriptor: | 2-dehydro-3-deoxygluconokinase, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a sugar kinase (Target EFI-502132) from Oceanicola granulosus with bound AMP, crystal form II
To be Published
|
|
4ECE
 
 | | Crystal structure of purine nucleoside phosphorylase (W16Y, W94Y, W178Y, H257W) mutant from human complexed with guanine | | Descriptor: | GUANINE, Purine nucleoside phosphorylase | | Authors: | Haapalainen, A.M, Ho, M.C, Suarez, J.J, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-03-26 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalytic Site Conformations in Human PNP by (19)F-NMR and Crystallography.
Chem.Biol., 20, 2013
|
|
3GPI
 
 | | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase | | Authors: | Ramagopal, U.A, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-23 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus
To be published
|
|
