7ROD
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 8 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROE
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 9 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROI
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 12 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7RO2
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 1 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROB
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 6 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7RO6
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 5 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROH
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 11 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROG
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 10 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROC
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 7 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7RO3
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 2 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7RO5
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 4 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
8DHW
 
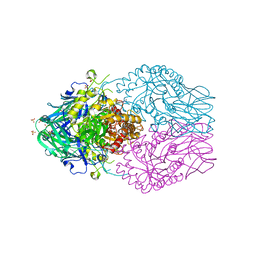 | | Treponema lecithinolyticum beta-glucuronidase in complex with a UNC4917-glucuronide conjugate | | Descriptor: | 4-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-2,7-bis(methylamino)pyrido[3',2':4,5]thieno[3,2-d]pyrimidine, Glycosyl hydrolase family 2, TIM barrel domain protein, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-06-28 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8DHL
 
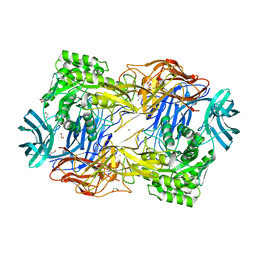 | | Tannerella forsythia beta-glucuronidase (L2) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycosyl hydrolase family 2, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-06-27 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8DHV
 
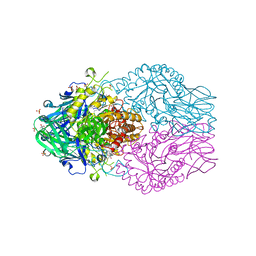 | | Treponema lecithinolyticum beta-glucuronidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Glycosyl hydrolase family 2, ... | | Authors: | Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2022-06-28 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Microbial beta-glucuronidases drive human periodontal disease etiology.
Sci Adv, 9, 2023
|
|
8DHE
 
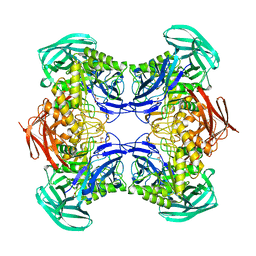 | |
5Z1V
 
 | | Crystal structure of AvrPib | | Descriptor: | AvrPib protein | | Authors: | Zhang, X, He, D, Zhao, Y.X, Taylor, I.A, Peng, Y.L, Yang, J, Liu, J.F. | | Deposit date: | 2017-12-28 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | A positive-charged patch and stabilized hydrophobic core are essential for avirulence function of AvrPib in the rice blast fungus.
Plant J., 96, 2018
|
|
7R1O
 
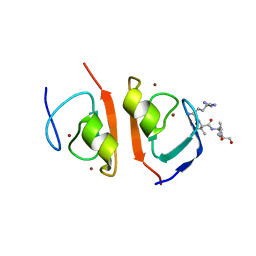 | | p62-ZZ domain of the human sequestosome in complex with dusquetide | | Descriptor: | Dusquetide, Sequestosome-1, ZINC ION | | Authors: | Hakansson, M, Hansson, M, Logan, D.T, Rozek, A, Donini, O. | | Deposit date: | 2022-02-03 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Dusquetide modulates innate immune response through binding to p62.
Structure, 30, 2022
|
|
8WUY
 
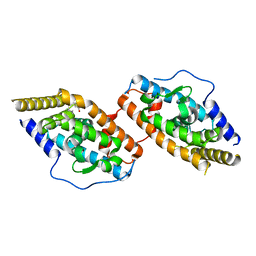 | | Crystal Structure of TR3 LBD in complex with para-positioned 3,4,5-trisubstituted benzene derivatives | | Descriptor: | Nuclear receptor subfamily 4immunitygroup A member 1, ~{N}-methyl-~{N}-octyl-3,4,5-tris(oxidanyl)benzamide | | Authors: | Hong, W.B, Chen, X.Q, Lin, T.W. | | Deposit date: | 2023-10-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and synthesis of anti-fibrotic compounds derived from para-positioned 3,4,5-trisubstituted benzene.
Bioorg.Chem., 144, 2024
|
|
2RHS
 
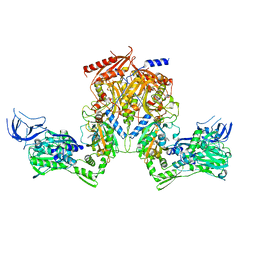 | | PheRS from Staphylococcus haemolyticus- rational protein engineering and inhibitor studies | | Descriptor: | 1-{3-[(4-pyridin-2-ylpiperazin-1-yl)sulfonyl]phenyl}-3-(1,3-thiazol-2-yl)urea, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Evdokimov, A.G, Mekel, M. | | Deposit date: | 2007-10-09 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational protein engineering in action: the first crystal structure of a phenylalanine tRNA synthetase from Staphylococcus haemolyticus.
J.Struct.Biol., 162, 2008
|
|
8AXT
 
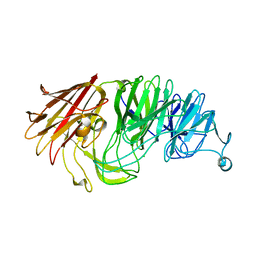 | | Sialidases and Fucosidases of Akkermansia muciniphila are key for rapid growth on colonic mucin and nutrient sharing amongst mucin-associated human gut microbiota | | Descriptor: | CALCIUM ION, CHLORIDE ION, Sialidase domain-containing protein | | Authors: | Sakanaka, H, Nielsen, T.S, Pichler, M.J, Nordberg Karlsson, E, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2022-08-31 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria.
Nat Commun, 14, 2023
|
|
8AYR
 
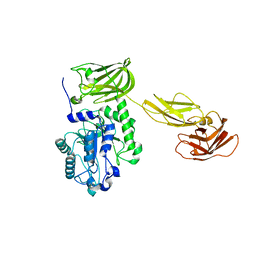 | | Sialidases and Fucosidases of Akkermansia muciniphila are key for rapid growth on colonic mucin and nutrient sharing amongst mucin-associated human gut microbiota | | Descriptor: | CALCIUM ION, Coagulation factor 5/8 type domain protein | | Authors: | Sakanaka, H, Nielsen, T.S, Pichler, M.J, Nordberg Karlsson, E, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria.
Nat Commun, 14, 2023
|
|
2RHQ
 
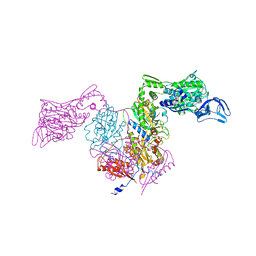 | | PheRS from Staphylococcus haemolyticus- rational protein engineering and inhibitor studies | | Descriptor: | 1-{3-[(4-pyridin-2-ylpiperazin-1-yl)sulfonyl]phenyl}-3-(1,3-thiazol-2-yl)urea, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Evdokimov, A.G, Mekel, M. | | Deposit date: | 2007-10-09 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational protein engineering in action: the first crystal structure of a phenylalanine tRNA synthetase from Staphylococcus haemolyticus.
J.Struct.Biol., 162, 2008
|
|
1XHS
 
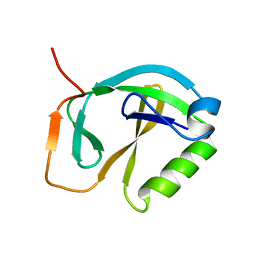 | | Solution NMR Structure of Protein ytfP from Escherichia coli. Northeast Structural Genomics Consortium Target ER111. | | Descriptor: | Hypothetical UPF0131 protein ytfP | | Authors: | Aramini, J.M, Huang, Y.J, Swapna, G.V.T, Paranji, R.K, Xiao, R, Shastry, R, Acton, T.B, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-09-20 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of Escherichia coli ytfP expands the structural coverage of the UPF0131 protein domain family.
Proteins, 68, 2007
|
|
8AXS
 
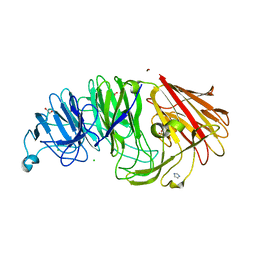 | | Sialidases and Fucosidases of Akkermansia muciniphila are key for rapid growth on colonic mucin and nutrient sharing amongst mucin-associated human gut microbiota | | Descriptor: | 1,2-ETHANEDIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Sakanaka, H, Nielsen, T.S, Pichler, M.J, Nordberg Karlsson, E, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2022-08-31 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria.
Nat Commun, 14, 2023
|
|
8AXI
 
 | | Sialidases and Fucosidases of Akkermansia muciniphila are key for rapid growth on colonic mucin and nutrient sharing amongst mucin-associated human gut microbiota | | Descriptor: | 1,2-ETHANEDIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Sakanaka, H, Nielsen, T.S, Pichler, M.J, Nordberg Karlsson, E, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2022-08-31 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria.
Nat Commun, 14, 2023
|
|
