4G9Q
 
 | | Crystal structure of a 4-carboxymuconolactone decarboxylase | | Descriptor: | 4-carboxymuconolactone decarboxylase | | Authors: | Hickey, H.D, Mcgillick, B.E, Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-24 | | Release date: | 2012-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a 4-carboxymuconolactone decarboxylase
To be Published
|
|
4GGH
 
 | | Crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, hepes, and ethylene glycol bound (ordered loops, space group c2221) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-06 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, hepes, and ethylene glycol bound (ordered loops, space group c2221)
To be Published
|
|
3I4S
 
 | | CRYSTAL STRUCTURE OF HISTIDINE TRIAD PROTEIN blr8122 FROM Bradyrhizobium japonicum | | Descriptor: | GLYCEROL, HISTIDINE TRIAD PROTEIN | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Freeman, J, Do, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-02 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | CRYSTAL STRUCTURE OF HISTIDINE TRIAD PROTEIN FROM Bradyrhizobium japonicum
To be Published
|
|
4GI5
 
 | | Crystal Structure Of a Putative quinone reductase from Klebsiella pneumoniae (Target PSI-013613) | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kumar, P.R, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Washington, E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a quinone reductase from Klebsiella pneumoniae with bound FAD
to be published
|
|
3I83
 
 | | Crystal structure of 2-dehydropantoate 2-reductase from Methylococcus capsulatus | | Descriptor: | 2-dehydropantoate 2-reductase, ACETIC ACID | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Chang, S, Sampathkumar, P, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of 2-dehydropantoate 2-reductase from Methylococcus capsulatus
To be Published
|
|
3I9X
 
 | | Crystal structure of a mutT/nudix family protein from Listeria innocua | | Descriptor: | GLYCEROL, mutT/nudix family protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Miller, S, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-13 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a mutT/nudix family protein from Listeria innocua
To be Published
|
|
3IMH
 
 | | CRYSTAL STRUCTURE OF GALACTOSE 1-EPIMERASE FROM Lactobacillus acidophilus NCFM | | Descriptor: | CHLORIDE ION, GLYCEROL, Galactose-1-epimerase, ... | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-10 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | CRYSTAL STRUCTURE OF GALACTOSE 1-EPIMERASE FROM Lactobacillus acidophilus
To be Published
|
|
3I9F
 
 | | Crystal structure of a putative type 11 methyltransferase from Sulfolobus solfataricus | | Descriptor: | Putative type 11 methyltransferase, ZINC ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Chang, S, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-10 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative type 11 methyltransferase from Sulfolobus solfataricus
To be Published
|
|
4FX6
 
 | | Crystal structure of the mutant V182A.R203A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-07-02 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3I42
 
 | |
4G9H
 
 | | Crystal structure of glutahtione s-transferase homolog from yersinia pestis, target EFI-501894, with bound glutathione | | Descriptor: | GLUTATHIONE, GLYCEROL, Glutathione S-transferase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glutahtione s-transferase homolog from yersinia pestis, target efi-501894, with bound glutathione
To be Published
|
|
4GC4
 
 | | Crystal structure of the mutant R160A.R203A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-07-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4204 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3I6V
 
 | | Crystal structure of a periplasmic His/Glu/Gln/Arg/opine family-binding protein from Silicibacter pomeroyi in complex with lysine | | Descriptor: | GLYCEROL, LYSINE, SODIUM ION, ... | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a periplasmic His/Glu/Gln/Arg/opine family-binding protein from Silicibacter pomeroyi in complex with lysine
To be Published
|
|
4FGS
 
 | | Crystal structure of a probable dehydrogenase protein | | Descriptor: | Probable dehydrogenase protein, SULFATE ION | | Authors: | Eswaramoorthy, S, Rice, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-06-04 | | Release date: | 2012-08-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a probable dehydrogenase protein
To be Published
|
|
3G1Y
 
 | | Crystal structure of the mutant D70N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with sulfate | | Descriptor: | Orotidine 5'-phosphate decarboxylase, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Chan, K.K, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: evidence for substrate destabilization.
Biochemistry, 48, 2009
|
|
3GHF
 
 | | Crystal structure of the septum site-determining protein minC from Salmonella typhimurium | | Descriptor: | CITRIC ACID, Septum site-determining protein minC | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the septum site-determining protein minC from Salmonella typhimurium
To be Published
|
|
4G56
 
 | | Crystal Structure of full length PRMT5/MEP50 complexes from Xenopus laevis | | Descriptor: | Hsl7 protein, MGC81050 protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Ho, M, Wilczek, C, Bonanno, J, Shechter, D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity
Plos One, 8, 2013
|
|
3GD6
 
 | | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with phosphate | | Descriptor: | Muconate cycloisomerase, PHOSPHATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-23 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with phosphate.
To be Published
|
|
4G9B
 
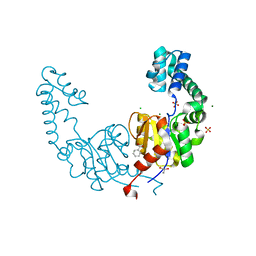 | | Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid | | Descriptor: | Beta-phosphoglucomutase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of beta-phosphoglucomutase homolog from escherichia coli, target efi-501172, with bound mg, open lid
To be Published
|
|
3GG7
 
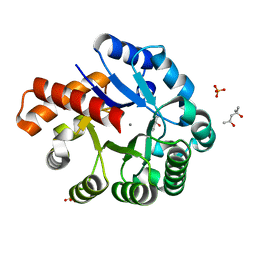 | | Crystal structure of an uncharacterized metalloprotein from Deinococcus radiodurans | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, MANGANESE (II) ION, ... | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Tang, B.K, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized metalloprotein from Deinococcus radiodurans
To be Published
|
|
3G8R
 
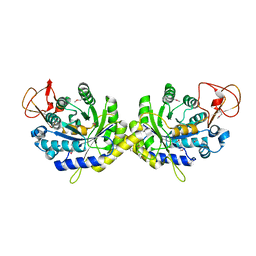 | | Crystal structure of putative spore coat polysaccharide biosynthesis protein E from Chromobacterium violaceum ATCC 12472 | | Descriptor: | Probable spore coat polysaccharide biosynthesis protein E, ZINC ION | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-02-12 | | Release date: | 2009-04-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of putative spore coat polysaccharide biosynthesis protein E from Chromobacterium violaceum ATCC 12472
To be Published
|
|
4GGB
 
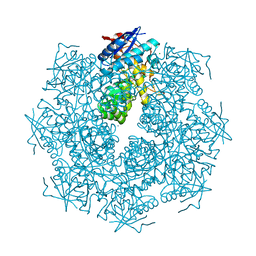 | | Crystal structure of a proposed galactarolactone cycloisomerase from agrobacterium tumefaciens, TARGET EFI-500704, WITH BOUND CA, DISORDERED LOOPS | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative uncharacterized protein | | Authors: | Vetting, M.W, Bouvier, J.T, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-06 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Purification, crystallization and structural elucidation of D-galactaro-1,4-lactone cycloisomerase from Agrobacterium tumefaciens involved in pectin degradation.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4GIR
 
 | | Crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, ethylene glycol and sulfate bound (ordered loops, space group P41212) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Enolase, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of an enolase family member from vibrio harveyi (efi-target 501692) with homology to mannonate dehydratase, with mg, ethylene glycol and sulfate bound (ordered loops, space group P41212)
To be Published
|
|
4GME
 
 | | Crystal structure of mannonate dehydratase (target EFI-502209) from caulobacter crescentus cb15 complexed with magnesium and d-mannonate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-MANNONIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-15 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mannonate Dehydratase from Caulobacter Crescentus Cb15
To be Published
|
|
3IJL
 
 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Pro-D-Glu; nonproductive substrate binding. | | Descriptor: | D-GLUTAMIC ACID, MAGNESIUM ION, Muconate cycloisomerase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
