3T69
 
 | | Crystal structure of a putative 2-dehydro-3-deoxygalactonokinase protein from Sinorhizobium meliloti | | Descriptor: | Putative 2-dehydro-3-deoxygalactonokinase, SULFATE ION | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-28 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of a putative 2-dehydro-3-deoxygalactonokinase protein from Sinorhizobium meliloti
To be Published
|
|
1I4Q
 
 | |
3T9P
 
 | | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme family protein from Roseovarius | | Descriptor: | FORMIC ACID, GLYCEROL, Mandelate racemase/muconate lactonizing enzyme family protein, ... | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-03 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme family protein from Roseovarius
To be Published
|
|
3T61
 
 | | Crystal Structure of a gluconokinase from Sinorhizobium meliloti 1021 | | Descriptor: | Gluconokinase, PHOSPHATE ION | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-28 | | Release date: | 2011-08-17 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a gluconokinase from Sinorhizobium meliloti 1021
To be Published
|
|
2HZT
 
 | | Crystal Structure of a putative HTH-type transcriptional regulator ytcD | | Descriptor: | Putative HTH-type transcriptional regulator ytcD | | Authors: | Madegowda, M, Eswaramoorthy, S, Desigan, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-09 | | Release date: | 2006-08-29 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a putative HTH-type transcription regulator ytcD
To be Published
|
|
2I5H
 
 | |
3C4R
 
 | | Crystal structure of an uncharacterized protein encoded by cryptic prophage | | Descriptor: | Uncharacterized protein | | Authors: | Sugadev, R, Burley, S.K, Swaminathan, S, Ozyurt, S, Luz, J, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-19 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an uncharacterized protein encoded by cryptic prophage.
To be Published
|
|
3T8L
 
 | | Crystal Structure of adenine deaminase with Mn/Fe | | Descriptor: | Adenine deaminase 2, UNKNOWN ATOM OR ION | | Authors: | Bagaria, A, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-08-01 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The catalase activity of diiron adenine deaminase.
Protein Sci., 20, 2011
|
|
5Z1I
 
 | | Crystal structure of the protozoal cytoplasmic ribosomal decoding site in complex with 6'-fluoro sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(fluoromethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(P*GP*CP*GP*UP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kanazawa, H, Hanessian, S, Kondo, J. | | Deposit date: | 2017-12-26 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structure-Based Design of a Eukaryote-Selective Antiprotozoal Fluorinated Aminoglycoside.
ChemMedChem, 13, 2018
|
|
2GU1
 
 | | Crystal structure of a zinc containing peptidase from vibrio cholerae | | Descriptor: | SODIUM ION, ZINC ION, Zinc peptidase | | Authors: | Sugadev, R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-28 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a putative lysostaphin peptidase from Vibrio cholerae.
Proteins, 72, 2008
|
|
5H1S
 
 | | Structure of the large subunit of the chloro-ribosome | | Descriptor: | 23S rRNA, 50S ribosomal protein L15, 50S ribosomal protein L17, ... | | Authors: | Ahmed, T, Yin, Z, Bhushan, S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-02-01 | | Last modified: | 2018-06-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the large subunit of the spinach chloroplast ribosome.
Sci Rep, 6, 2016
|
|
2KSS
 
 | | NMR structure of Myxococcus xanthus antirepressor CarS1 | | Descriptor: | Carotenogenesis protein carS | | Authors: | Jimenez, M, Gonzalez, C, Padmanabhan, S, Leon, E, Navarro-Aviles, G, Elias-Arnanz, M. | | Deposit date: | 2010-01-13 | | Release date: | 2010-05-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A bacterial antirepressor with SH3 domain topology mimics operator DNA in sequestering the repressor DNA recognition helix.
Nucleic Acids Res., 38, 2010
|
|
3TFO
 
 | | Crystal structure of a putative 3-oxoacyl-(acyl-carrier-protein) reductase from Sinorhizobium meliloti | | Descriptor: | HEXANE-1,6-DIOL, SULFATE ION, putative 3-oxoacyl-(acyl-carrier-protein) reductase | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-16 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-(acyl-carrier-protein) reductase from Sinorhizobium meliloti
To be Published
|
|
3T66
 
 | | Crystal structure of Nickel ABC transporter from Bacillus halodurans | | Descriptor: | CALCIUM ION, Nickel ABC transporter (Nickel-binding protein) | | Authors: | Agarwal, R, Bonanno, J.B, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-28 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Nickel ABC transporter from Bacillus halodurans
To be Published
|
|
3TFX
 
 | | Crystal structure of Orotidine 5'-phosphate decarboxylase from Lactobacillus acidophilus | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Satyanarayana, L, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-16 | | Release date: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of Orotidine 5'-phosphate decarboxylase from Lactobacillus acidophilus
To be Published
|
|
2GVC
 
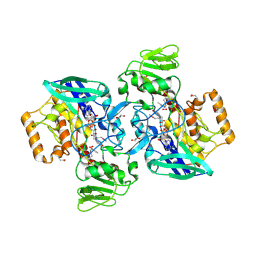 | | Crystal structure of flavin-containing monooxygenase (FMO)from S.pombe and substrate (methimazole) complex | | Descriptor: | 1-METHYL-1,3-DIHYDRO-2H-IMIDAZOLE-2-THIONE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Mechanism of action of a flavin-containing monooxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1I1E
 
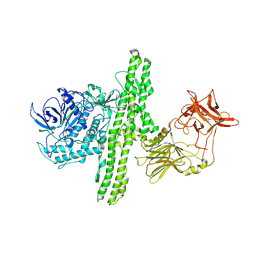 | | CRYSTAL STRUCTURE OF CLOSTRIDIUM BOTULINUM NEUROTOXIN B COMPLEXED WITH DOXORUBICIN | | Descriptor: | BOTULINUM NEUROTOXIN TYPE B, DOXORUBICIN, SULFATE ION, ... | | Authors: | Eswaramoorthy, S, Kumaran, D, Swaminathan, S. | | Deposit date: | 2001-02-01 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic evidence for doxorubicin binding to the receptor-binding site in Clostridium botulinum neurotoxin B.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1I4R
 
 | |
3ROS
 
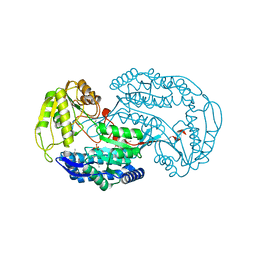 | |
2GV8
 
 | | Crystal structure of flavin-containing monooxygenase (FMO) from S.pombe and NADPH cofactor complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of action of a flavin-containing monooxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5Z1H
 
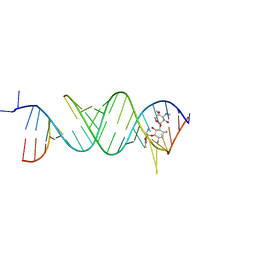 | | Crystal structure of the bacterial ribosomal decoding site in complex with 6'-fluoro sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(fluoromethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kanazawa, H, Hanessian, S, Kondo, J. | | Deposit date: | 2017-12-26 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of a Eukaryote-Selective Antiprotozoal Fluorinated Aminoglycoside.
ChemMedChem, 13, 2018
|
|
1Z0H
 
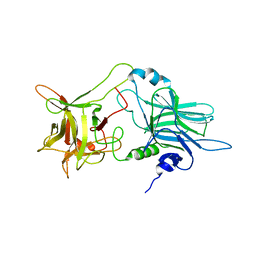 | | N-terminal helix reorients in recombinant C-fragment of Clostridium botulinum type B | | Descriptor: | Botulinum neurotoxin type B | | Authors: | Jayaraman, S, Eswarmoorthy, S, Ashraf, S.A, Smith, L.A, Swaminathan, S. | | Deposit date: | 2005-03-01 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-terminal helix reorients in recombinant C-fragment of Clostridium botulinum type B.
Biochem.Biophys.Res.Commun., 330, 2005
|
|
3TPC
 
 | | Crystal structure of a hypothtical protein SMa1452 from Sinorhizobium meliloti 1021 | | Descriptor: | Short chain alcohol dehydrogenase-related dehydrogenase | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-09-07 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of a hypothtical protein SMa1452 from Sinorhizobium meliloti 1021
To be Published
|
|
1DPE
 
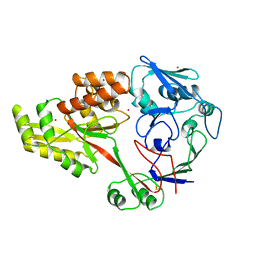 | | DIPEPTIDE-BINDING PROTEIN | | Descriptor: | CADMIUM ION, DIPEPTIDE-BINDING PROTEIN | | Authors: | Nickitenko, A.V, Trakhanov, S, Quiocho, F.A. | | Deposit date: | 1995-07-25 | | Release date: | 1996-08-17 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2 A resolution structure of DppA, a periplasmic dipeptide transport/chemosensory receptor.
Biochemistry, 34, 1995
|
|
4HUJ
 
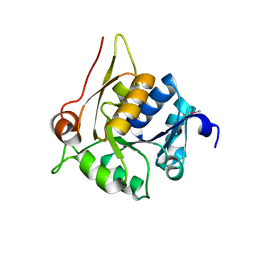 | | Crystal structure of a hypothetical protein SMa0349 from Sinorhizobium meliloti | | Descriptor: | Uncharacterized protein | | Authors: | Rice, S, Eswaramoorthy, S, Chamala, S, Evans, B, Foti, F, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-11-02 | | Release date: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of a hypothetical protein SMa0349 from Sinorhizobium meliloti
To be Published
|
|
