1Q6C
 
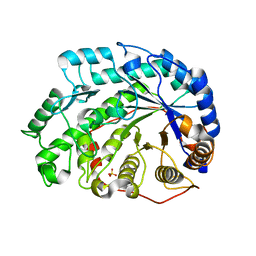 | | Crystal Structure of Soybean Beta-Amylase Complexed with Maltose | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-amylase | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
5VEN
 
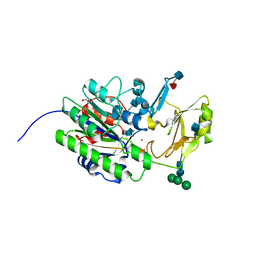 | | Murine ectonucleotide pyrophosphatase / phosphodiesterase 5 (ENPP5, NPP5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ectonucleotide pyrophosphatase/phosphodiesterase family member 5, ... | | Authors: | Gorelik, A, Randriamihaja, A, Illes, K, Nagar, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A key tyrosine substitution restricts nucleotide hydrolysis by the ectoenzyme NPP5.
FEBS J., 284, 2017
|
|
1JH6
 
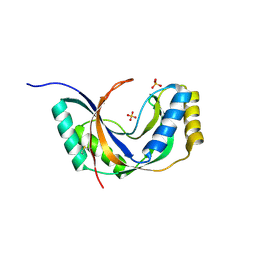 | | Semi-reduced Cyclic Nucleotide Phosphodiesterase from Arabidopsis thaliana | | Descriptor: | SULFATE ION, cyclic phosphodiesterase | | Authors: | Hofmann, A, Grella, M, Botos, I, Filipowicz, W, Wlodawer, A. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the semireduced and inhibitor-bound forms of cyclic nucleotide phosphodiesterase from Arabidopsis thaliana.
J.Biol.Chem., 277, 2002
|
|
1QBB
 
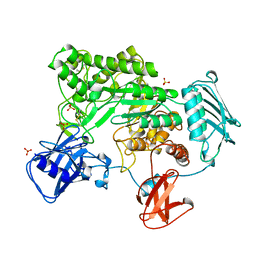 | | BACTERIAL CHITOBIASE COMPLEXED WITH CHITOBIOSE (DINAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-07 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1JNJ
 
 | | NMR solution structure of the human beta2-microglobulin | | Descriptor: | beta2-microglobulin | | Authors: | Verdone, G, Corazza, A, Viglino, P, Pettirossi, F, Giorgetti, S, Mangione, P, Andreola, A, Stoppini, M, Bellotti, V, Esposito, G. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human beta2-microglobulin reveals the prodromes of its amyloid transition.
Protein Sci., 11, 2002
|
|
7BXV
 
 | | 11A1 antibody-peptide complex | | Descriptor: | 1,2-ETHANEDIOL, Fab of the 11A1 antibody H chain, Fab of the 11A1 antibody L chain, ... | | Authors: | Irie, K, Irie, Y, Kita, A, Miki, K. | | Deposit date: | 2020-04-21 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | APOE epsilon 4 allele advances the age-dependent decline of amyloid beta clearance in the human cortex.
Biorxiv, 2022
|
|
4JFL
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one | | Descriptor: | 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4D79
 
 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA, in complex with ATP at 1.768 Angstroem resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, POTASSIUM ION, ... | | Authors: | Lopez-Estepa, M, Arda, A, Savko, M, Round, A, Shepard, W, Bruix, M, Coll, M, Fernandez, F.J, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2014-11-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | The Crystal Structure and Small-Angle X-Ray Analysis of Csdl/Tcda Reveal a New tRNA Binding Motif in the Moeb/E1 Superfamily.
Plos One, 10, 2015
|
|
4AFU
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, FYNOMER | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
8U6X
 
 | | ATP-dependent DNA ligase Lig E from Neisseria gonorrhoeae | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*GP*CP*GP*AP*CP*CP*CP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3'), ... | | Authors: | Williamson, A, Pan, J. | | Deposit date: | 2023-09-14 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A role for the ATP-dependent DNA ligase lig E of Neisseria gonorrhoeae in biofilm formation.
Bmc Microbiol., 24, 2024
|
|
2EVV
 
 | | Crystal Structure of the PEBP-like Protein of Unknown Function HP0218 from Helicobacter pylori | | Descriptor: | GLYCEROL, SULFATE ION, hypothetical protein HP0218 | | Authors: | Kim, Y, Xu, X, Hong, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-11-01 | | Release date: | 2005-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of the Hypothetical Protein HP0218 from Helicobacter pylori
To be Published
|
|
4D7A
 
 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA, in complex with AMP at 1.801 Angstroem resolution | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lopez-Estepa, M, Arda, A, Savko, M, Round, A, Shepard, W, Bruix, M, Coll, M, Fernandez, F.J, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2014-11-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The Crystal Structure and Small-Angle X-Ray Analysis of Csdl/Tcda Reveal a New tRNA Binding Motif in the Moeb/E1 Superfamily.
Plos One, 10, 2015
|
|
6BV8
 
 | | Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana complexed with Mn after 3-hour soak with juglone | | Descriptor: | 5-hydroxynaphthalene-1,4-dione, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Karasik, A, Wu, N, Fierke, C.A, Koutmos, M. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of protein-only RNase P with Gambogic acid and Juglone
To Be Published
|
|
4ZGR
 
 | | Structural studies on a non-toxic homologue of type II RIPs from Momordica charantia (bitter gourd) in complex with T-Antigen. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Chandran, T, Sharma, A, Vijayan, M. | | Deposit date: | 2015-04-23 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural studies on a non-toxic homologue of type II RIPs from bitter gourd: Molecular basis of non-toxicity, conformational selection and glycan structure.
J.Biosci., 40, 2015
|
|
1DD5
 
 | | CRYSTAL STRUCTURE OF THERMOTOGA MARITIMA RIBOSOME RECYCLING FACTOR, RRF | | Descriptor: | ACETIC ACID, RIBOSOME RECYCLING FACTOR | | Authors: | Selmer, M, Al-Karadaghi, S, Hirokawa, G, Kaji, A, Liljas, A. | | Deposit date: | 1999-11-08 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic.
Science, 286, 1999
|
|
3LN4
 
 | | Crystal structure of HLA-B*4103 in complex with a 16mer self-peptide derived from heterogeneous nuclear ribonucleoproteins C1/C2 | | Descriptor: | 16-mer peptide from Heterogeneous nuclear ribonucleoproteins C1/C2, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Theodossis, A, Gras, S, Rossjohn, J. | | Deposit date: | 2010-02-01 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | The impact of human leukocyte antigen (HLA) micropolymorphism on ligand specificity within the HLA-B*41 allotypic family
Haematologica, 96, 2011
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
1P2A
 
 | | The structure of cyclin dependent kinase 2 (CKD2) with a trisubstituted naphthostyril inhibitor | | Descriptor: | 5-[(2-AMINOETHYL)AMINO]-6-FLUORO-3-(1H-PYRROL-2-YL)BENZO[CD]INDOL-2(1H)-ONE, Cell division protein kinase 2 | | Authors: | Liu, J.-J, Dermatakis, A, Lukacs, C.M, Konzelmann, F, Chen, Y, Kammlott, U, Depinto, W, Yang, H, Yin, X, Chen, Y, Schutt, A, Simcox, M.E, Luk, K.-C. | | Deposit date: | 2003-04-15 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3,5,6-Trisubstituted Naphthostyrils as CDK2 Inhibitors
BIOORG.MED.CHEM., 13, 2003
|
|
3LBL
 
 | | Structure of human MDM2 protein in complex with Mi-63-analog | | Descriptor: | (2'R,3R,4'R,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(2-morpholin-4-ylethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
1T6C
 
 | | Structural characterization of the Ppx/GppA protein family: crystal structure of the Aquifex aeolicus family member | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kristensen, O, Laurberg, M, Liljas, A, Kastrup, J.S, Gajhede, M. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural characterization of the stringent response related exopolyphosphatase/guanosine pentaphosphate phosphohydrolase protein family
Biochemistry, 43, 2004
|
|
6GQE
 
 | |
2B6H
 
 | | Structure of human ADP-ribosylation factor 5 | | Descriptor: | ADP-ribosylation factor 5, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tempel, W, Atanassova, A, Sundarajan, E, Dimov, S, Shehab, I, Lew, J, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-10-01 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Structure of human ADP-ribosylation factor 5
To be Published
|
|
1QBA
 
 | | BACTERIAL CHITOBIASE, GLYCOSYL HYDROLASE FAMILY 20 | | Descriptor: | CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
5VEM
 
 | | Human ectonucleotide pyrophosphatase / phosphodiesterase 5 (ENPP5, NPP5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ectonucleotide pyrophosphatase/phosphodiesterase family member 5, ... | | Authors: | Gorelik, A, Randriamihaja, A, Illes, K, Nagar, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A key tyrosine substitution restricts nucleotide hydrolysis by the ectoenzyme NPP5.
FEBS J., 284, 2017
|
|
2PEG
 
 | | Crystal structure of Trematomus bernacchii hemoglobin in a partial hemichrome state | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vergara, A, Franzese, M, Merlino, A, Vitagliano, L, Mazzarella, L. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural characterization of ferric hemoglobins from three antarctic fish species of the suborder notothenioidei.
Biophys.J., 93, 2007
|
|
