1ZPV
 
 | | ACT domain protein from Streptococcus pneumoniae | | Descriptor: | ACT domain protein, POTASSIUM ION | | Authors: | Osipiuk, J, Hatzos, C, Abdullah, J, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-17 | | Release date: | 2005-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of ACT domain protein from Streptococcus pneumoniae
To be Published
|
|
1ZTS
 
 | | Solution Structure of Bacillus Subtilis Protein YQBG: Northeast Structural Genomics Consortium Target SR215 | | Descriptor: | Hypothetical protein yqbG | | Authors: | Liu, G, Shen, Y, Xiao, R, Acton, T, Ma, L, Joachimiak, A, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-27 | | Release date: | 2005-07-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein yqbG from Bacillus subtilis reveals a novel alpha-helical protein fold.
Proteins, 62, 2006
|
|
1YQG
 
 | | Crystal structure of a pyrroline-5-carboxylate reductase from neisseria meningitides mc58 | | Descriptor: | SULFATE ION, pyrroline-5-carboxylate reductase | | Authors: | Chang, C, Joachimiak, A, Li, H, Collart, F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-01 | | Release date: | 2005-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Delta(1)-Pyrroline-5-carboxylate Reductase from Human Pathogens Neisseria meningitides and Streptococcus pyogenes
J.Mol.Biol., 354, 2005
|
|
4ICH
 
 | | Crystal structure of a putative TetR family transcriptional regulator from Saccharomonospora viridis DSM 43017 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-02 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a putative TetR family transcriptional regulator from Saccharomonospora viridis DSM 43017
To be Published
|
|
2BE3
 
 | |
4J41
 
 | | The crystal structure of a secreted protein EsxB (Mutant P67A) from Bacillus anthracis str. Sterne | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-06 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.522 Å) | | Cite: | The crystal structure of a secreted protein EsxB (Mutant P67A) from Bacillus anthracis str. Sterne
To be Published
|
|
4ONW
 
 | | Crystal structure of the catalytic domain of DapE protein from V.cholerea | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, ACETATE ION, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Jedrzejczak, R, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-29 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Dimerization Domain in DapE Enzymes Is required for Catalysis.
Plos One, 9, 2014
|
|
4ISC
 
 | | Crystal structure of a putative Methyltransferase from Pseudomonas syringae | | Descriptor: | BETA-MERCAPTOETHANOL, Methyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of a putative Methyltransferase from Pseudomonas syringae
To be Published
|
|
4ISX
 
 | | The crystal structure of maltose o-acetyltransferase from clostridium difficile 630 in complex with acetyl-coa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETYL COENZYME *A, Maltose O-acetyltransferase | | Authors: | Tan, K, Gu, G, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-17 | | Release date: | 2013-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | The crystal structure of maltose o-acetyltransferase from clostridium difficile 630 in complex with acetyl-coa
To be Published
|
|
4IYI
 
 | | The crystal structure of a secreted protein EsxB (wild-type, C-term. His-tagged) from Bacillus anthracis str. Sterne | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.083 Å) | | Cite: | The crystal structure of a secreted protein EsxB (wild-type, C-term. His-tagged) from Bacillus anthracis str. Sterne
To be Published
|
|
4OP4
 
 | | Crystal structure of the catalytic domain of DapE protein from V.cholerea in the Zn bound form | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, GLYCEROL, ... | | Authors: | Nocek, B, Makowska-Grzyska, M, Jedrzejczak, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | The Dimerization Domain in DapE Enzymes Is required for Catalysis.
Plos One, 9, 2014
|
|
4IYL
 
 | | 30S ribosomal protein S15 from Campylobacter jejuni | | Descriptor: | 30S ribosomal protein S15 | | Authors: | Osipiuk, J, Nocek, B, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | 30S ribosomal protein S15 from Campylobacter jejuni
To be Published
|
|
4IXH
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Cryptosporidium parvum | | Descriptor: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, INOSINIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Kavitha, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-25 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Optimization of Benzoxazole-Based Inhibitors of Cryptosporidium parvum Inosine 5'-Monophosphate Dehydrogenase.
J.Med.Chem., 56, 2013
|
|
4IYH
 
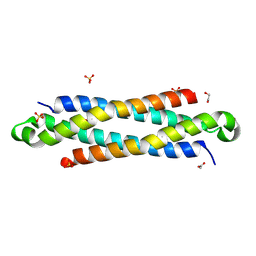 | | The crystal structure of a secreted protein EsxB (SeMet-labeled, C-term. His-Tagged) from Bacillus anthracis str. Sterne | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | The crystal structure of a secreted protein EsxB (SeMet-labeled, C-term. His-Tagged) from Bacillus anthracis str. Sterne
To be Published
|
|
4J42
 
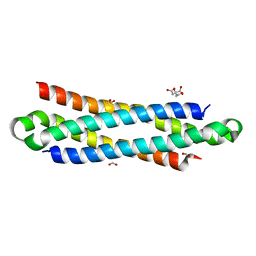 | | The crystal structure of a secreted protein EsxB (Mutant Y65F) from Bacillus anthracis str. Sterne | | Descriptor: | CITRATE ANION, FORMIC ACID, secreted protein EsxB | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-06 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The crystal structure of a secreted protein EsxB (Mutant Y65F) from Bacillus anthracis str. Sterne
To be Published
|
|
4O2I
 
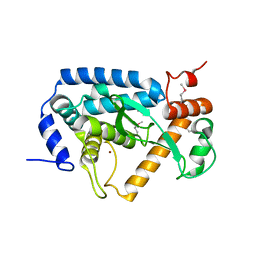 | | The crystal structure of non-LEE encoded type III effector C from Citrobacter rodentium | | Descriptor: | Non-LEE encoded type III effector C, ZINC ION | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of non-LEE encoded type III effector C from Citrobacter rodentium
To be Published
|
|
4IT5
 
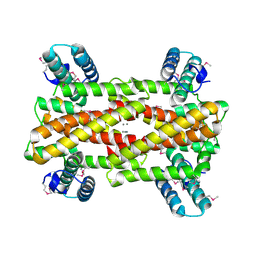 | | Chaperone HscB from Vibrio cholerae | | Descriptor: | CALCIUM ION, Co-chaperone protein HscB homolog | | Authors: | Osipiuk, J, Gu, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-17 | | Release date: | 2013-01-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Chaperone HscB from Vibrio cholerae.
To be Published
|
|
4JD1
 
 | | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames | | Descriptor: | FOSFOMYCIN, Metallothiol transferase FosB 2, TRIETHYLENE GLYCOL, ... | | Authors: | Maltseva, N, Kim, Y, Jedrzejczak, R, Sharma, S.V, Hamilton, C.J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-22 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames
To be Published
|
|
4JGR
 
 | | The crystal structure of sporulation kinase D mutant sensor domain, R131A, from Bacillus subtilis subsp at 2.4A resolution | | Descriptor: | ACETIC ACID, GLYCEROL, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4JWO
 
 | | The crystal structure of a possible phosphate binding protein from Planctomyces limnophilus DSM 3776 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Tan, K, Gu, M, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-27 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The crystal structure of a possible phosphate binding protein from Planctomyces limnophilus DSM 3776
To be Published
|
|
4O23
 
 | | Crystal structure of mono-zinc form of succinyl diaminopimelate desuccinylase from Neisseria meningitidis MC58 | | Descriptor: | SULFATE ION, Succinyl-diaminopimelate desuccinylase, ZINC ION | | Authors: | Nocek, B, Holz, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Inhibition of the dapE-Encoded N-Succinyl-L,L-diaminopimelic Acid Desuccinylase from Neisseria meningitidis by L-Captopril.
Biochemistry, 54, 2015
|
|
4NE4
 
 | | Crystal structure of ABC transporter substrate binding protein ProX from Agrobacterium tumefaciens cocrystalized with BTB | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ABC transporter, substrate binding protein (Proline/glycine/betaine), ... | | Authors: | Tkaczuk, K.L, Nicholls, R, Kagan, O, Chruszcz, M, Domagalski, M.J, Savchenko, A, Joachimiak, A, Murshudov, G, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-28 | | Release date: | 2013-11-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of ABC transporter substrate binding protein ProX from Agrobacterium tumefaciens cocrystalized with BTB
To be Published
|
|
4IPT
 
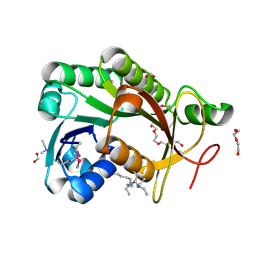 | | The crystal structure of a short-chain dehydrogenases/reductase (ethylated) from Veillonella parvula DSM 2008 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-10 | | Release date: | 2013-02-06 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | The crystal structure of a short-chain dehydrogenases/reductase (ethylated) from Veillonella parvula DSM 2008
To be Published
|
|
4IR0
 
 | | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames | | Descriptor: | 1,2-ETHANEDIOL, FOSFOMYCIN, Metallothiol transferase FosB 2, ... | | Authors: | Maltseva, N, Kim, Y, Jedrzejczak, R, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Metallothiol Transferase FosB 2 from Bacillus anthracis str. Ames
To be Published
|
|
1YZF
 
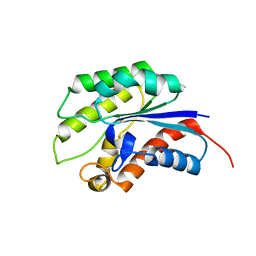 | | Crystal structure of the lipase/acylhydrolase from Enterococcus faecalis | | Descriptor: | lipase/acylhydrolase | | Authors: | Zhang, R, Hatzos, C, Clancy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-28 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the lipase/acylhydrolase from Enterococcus faecalis
To be Published
|
|
