8QE0
 
 | | Crystal structure of human MAT2a bound to S-Adenosylmethionine and Compound 12 | | Descriptor: | 4-[4-[bis(fluoranyl)methoxy]phenyl]-3-cyclopropyl-2~{H}-pyrazolo[4,3-b]pyridin-5-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Development of a Series of Pyrrolopyridone MAT2A Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QE2
 
 | | Crystal structure of human MAT2a bound to S-Adenosylmethionine and Compound 21 | | Descriptor: | 4-[4-[bis(fluoranyl)methoxy]phenyl]-3-cyclopropyl-6-(2-methylindazol-5-yl)-2~{H}-pyrazolo[4,3-b]pyridin-5-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.109 Å) | | Cite: | Development of a Series of Pyrrolopyridone MAT2A Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QE3
 
 | | Crystal structure of human MAT2a bound to S-Adenosylmethionine and Compound 31 | | Descriptor: | 3-cyclopropyl-6-(2-methylindazol-5-yl)-4-(6-methylpyridin-3-yl)-2~{H}-pyrazolo[4,3-b]pyridin-5-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.089 Å) | | Cite: | Development of a Series of Pyrrolopyridone MAT2A Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QE1
 
 | | Crystal structure of human MAT2a bound to S-Adenosylmethionine and Compound 15 | | Descriptor: | 4-[4-[bis(fluoranyl)methoxy]phenyl]-3-cyclopropyl-6-(4-methoxyphenyl)-2~{H}-pyrazolo[4,3-b]pyridin-5-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.095 Å) | | Cite: | Development of a Series of Pyrrolopyridone MAT2A Inhibitors.
J.Med.Chem., 67, 2024
|
|
8QDZ
 
 | |
8GZ9
 
 | | Cryo-EM structure of Abeta2 fibril polymorph2 | | Descriptor: | peptide self-assembled antimicrobial fibrils | | Authors: | Xia, W.C, Zhang, M.M, Liu, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Engineering of antimicrobial peptide fibrils with feedback degradation of bacterial-secreted enzymes.
Chem Sci, 14, 2023
|
|
8GZ8
 
 | | Cryo-EM structure of Abeta2 fibril polymorph1 | | Descriptor: | peptide self-assembled antimicrobial fibrils | | Authors: | Xia, W.C, Zhang, M.M, Liu, C. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Engineering of antimicrobial peptide fibrils with feedback degradation of bacterial-secreted enzymes.
Chem Sci, 14, 2023
|
|
8HOA
 
 | | ScRIPK mutant K124R | | Descriptor: | (2R,5R)-hexane-2,5-diol, ADENOSINE-5'-TRIPHOSPHATE, FORMIC ACID, ... | | Authors: | Fang, J.L, Zhang, M.Q, Ming, Z.H. | | Deposit date: | 2022-12-09 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.676 Å) | | Cite: | Receptor-like cytoplasmic kinase ScRIPK in sugarcane regulates disease resistance and drought tolerance in Arabidopsis.
Front Plant Sci, 14, 2023
|
|
8HO6
 
 | | ScRIPK WT | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ScRIPK kinase protein | | Authors: | Fang, J.L, Zhang, M.Q. | | Deposit date: | 2022-12-09 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Receptor-like cytoplasmic kinase ScRIPK in sugarcane regulates disease resistance and drought tolerance in Arabidopsis.
Front Plant Sci, 14, 2023
|
|
8HOD
 
 | | ScRIPK MUTANT-S253A, T254A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Hypothetical protein | | Authors: | Fang, J.L, Zhang, M.Q. | | Deposit date: | 2022-12-09 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Receptor-like cytoplasmic kinase ScRIPK in sugarcane regulates disease resistance and drought tolerance in Arabidopsis.
Front Plant Sci, 14, 2023
|
|
8QDY
 
 | |
6VSP
 
 | | Structure of Serratia marcescens 2,3-butanediol dehydrogenase mutant Q247A | | Descriptor: | 1,2-ETHANEDIOL, 2,3-butanediol dehydrogenase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phylogenetics-based identification and characterization of a superior 2,3-butanediol dehydrogenase for Zymomonas mobilis expression.
Biotechnol Biofuels, 13, 2020
|
|
6O8S
 
 | | Syn-safencin 56 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6O8T
 
 | | Syn-safencin 96 | | Descriptor: | Circular bacteriocin, circularin A/uberolysin family | | Authors: | Fields, F.R, Lee, S.W. | | Deposit date: | 2019-03-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Synthetic Antimicrobial Peptide Tuning Permits Membrane Disruption and Interpeptide Synergy.
Acs Pharmacol Transl Sci, 3, 2020
|
|
5H19
 
 | | EED in complex with PRC2 allosteric inhibitor EED162 | | Descriptor: | 5-(furan-2-ylmethylamino)-9-(phenylmethyl)-8,10-dihydro-7H-[1,2,4]triazolo[3,4-a][2,7]naphthyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
4YH1
 
 | |
4YGY
 
 | |
8I3U
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 14B1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 14B1, Light chain of Fab 14B1, ... | | Authors: | Li, Z, Yu, F, Cao, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
8I3S
 
 | | Local CryoEM structure of the SARS-CoV-2 S6P in complex with 7B3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain od Fab 7B3, Light chain of Fab 7B3, ... | | Authors: | Li, Z, Yu, F, Cao, S, ZHao, H. | | Deposit date: | 2023-01-17 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Broadly neutralizing antibodies derived from the earliest COVID-19 convalescents protect mice from SARS-CoV-2 variants challenge.
Signal Transduct Target Ther, 8, 2023
|
|
6V7A
 
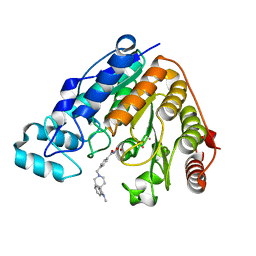 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with NF2657 | | Descriptor: | Hdac6 protein, N-hydroxy-4-[(1-methyl-1,2-dihydro-1'H-spiro[indole-3,4'-piperidin]-1'-yl)methyl]benzamide, POTASSIUM ION, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.0874176 Å) | | Cite: | Spiroindoline-Capped Selective HDAC6 Inhibitors: Design, Synthesis, Structural Analysis, and Biological Evaluation.
Acs Med.Chem.Lett., 11, 2020
|
|
6EZV
 
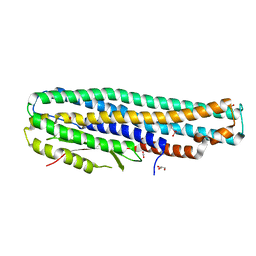 | | The cytotoxin MakA from Vibrio cholerae | | Descriptor: | ACETATE ION, CACODYLATE ION, GLYCEROL, ... | | Authors: | Persson, K, Dongre, M, Wai, S.N. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flagella-mediated secretion of a novelVibrio choleraecytotoxin affecting both vertebrate and invertebrate hosts.
Commun Biol, 1, 2018
|
|
7ZWA
 
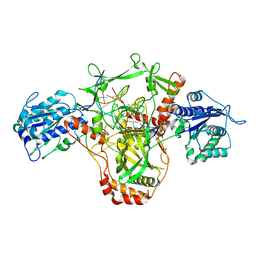 | | CryoEM structure of Ku heterodimer bound to DNA and PAXX | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*T)-3'), PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
7ZYG
 
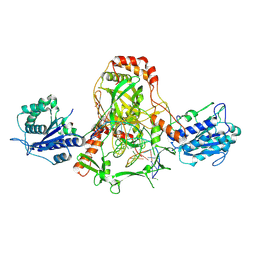 | | CryoEM structure of Ku heterodimer bound to DNA, PAXX and XLF | | Descriptor: | DNA, Non-homologous end-joining factor 1, PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
5H24
 
 | | EED in complex with PRC2 allosteric inhibitor compound 8 | | Descriptor: | 5-(furan-2-ylmethylamino)-[1,2,4]triazolo[4,3-a]pyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
6T3B
 
 | | Crystal structure of PI3Kgamma with a dihydropurinone inhibitor (compound 4) | | Descriptor: | 2-[(4-methoxy-2-methyl-phenyl)amino]-7-methyl-9-(4-oxidanylcyclohexyl)purin-8-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Petersen, J, Oster, L, Schimpl, M, Goldberg, F.W, Finlay, M.R.V, Ting, A.K.T, Beattie, D, Lamont, G.M, Fallan, C, Wrigley, G.L, Howard, M.R, Williamson, B, Davies, B.R, Cadogan, E.B, Ramos-Montoya, A, Dean, E. | | Deposit date: | 2019-10-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | The Discovery of 7-Methyl-2-[(7-methyl[1,2,4]triazolo[1,5-a]pyridin-6-yl)amino]-9-(tetrahydro-2H-pyran-4-yl)-7,9-dihydro-8H-purin-8-one (AZD7648), a Potent and Selective DNA-Dependent Protein Kinase (DNA-PK) Inhibitor.
J.Med.Chem., 63, 2020
|
|
