9ELI
 
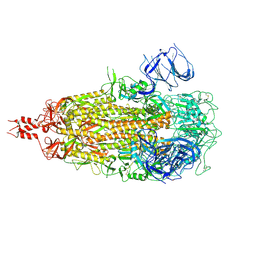 | | Cryo-EM structure of SARS-CoV-2 Omicron KP.3.1.1 spike protein (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Feng, Z, Huang, J, Ward, A.B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural and functional insights into the evolution of SARS-CoV-2 KP.3.1.1 spike protein.
Cell Rep, 44, 2025
|
|
5XRQ
 
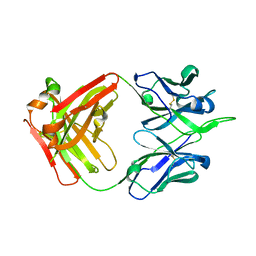 | | Crystal structure of human monoclonal antibody H3v-47 | | Descriptor: | Fab H3v-47 heavy chain, Fab H3v-47 light chain | | Authors: | Zhang, H, Willson, I.A. | | Deposit date: | 2017-06-09 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA.
Nat Commun, 9, 2018
|
|
3YPI
 
 | |
8D0Z
 
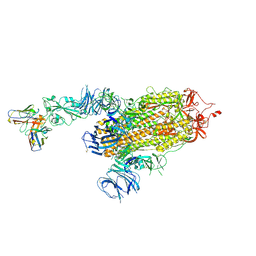 | | S728-1157 IgG in complex with SARS-CoV-2-6P-Mut7 Spike protein (focused refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S728-1157 Fab heavy chain variable region, S728-1157 Fab light chain variable region, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2022-05-26 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Site of vulnerability on SARS-CoV-2 spike induces broadly protective antibody against antigenically distinct Omicron subvariants.
J.Clin.Invest., 133, 2023
|
|
2OOK
 
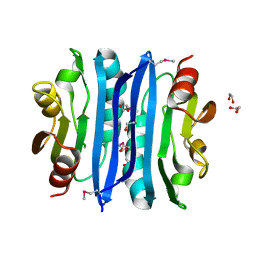 | |
2OOC
 
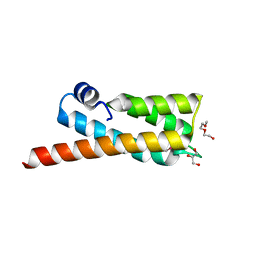 | |
2PV7
 
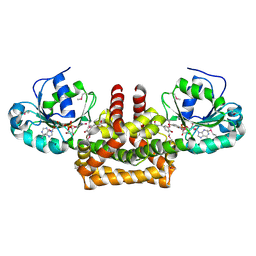 | |
2Q3L
 
 | |
2Q86
 
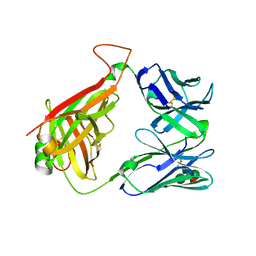 | | Structure of the mouse invariant NKT cell receptor Valpha14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Valpha14 TCR, Vbeta8.2, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2007-06-08 | | Release date: | 2008-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Mouse CD1d-iGb3 Complex and its Cognate Valpha14 T Cell Receptor Suggest a Model for Dual Recognition of Foreign and Self Glycolipids.
J.Mol.Biol., 377, 2008
|
|
2Q8U
 
 | |
2Q83
 
 | |
2QAD
 
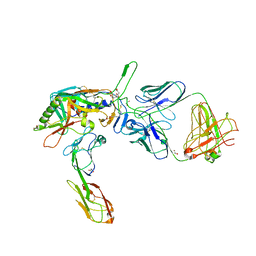 | | Structure of tyrosine-sulfated 412d antibody complexed with HIV-1 YU2 gp120 and CD4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Huang, C.-C, Tang, M, Robinson, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2007-06-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4
Science, 317, 2007
|
|
3HSA
 
 | |
3T2L
 
 | |
3SY6
 
 | |
3G23
 
 | |
3UP6
 
 | |
3UWS
 
 | |
3HN7
 
 | |
3H0N
 
 | |
3GO5
 
 | |
3HBZ
 
 | |
3IRB
 
 | |
3G0T
 
 | |
3TCL
 
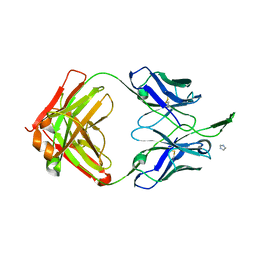 | | Crystal Structure of HIV-1 Neutralizing Antibody CH04 | | Descriptor: | CH04 Heavy Chain Fab, CH04 Light Chain Fab, IMIDAZOLE | | Authors: | Louder, R.K, McLellan, J.S, Pancera, M, Yang, Y, Zhang, B, Bonsignori, M, Kwong, P.D. | | Deposit date: | 2011-08-09 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
