3DEB
 
 | |
3LKI
 
 | |
3DD5
 
 | | Glomerella cingulata E600-cutinase complex | | Descriptor: | Cutinase, DIETHYL PHOSPHONATE | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-05 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3DCN
 
 | | Glomerella cingulata apo cutinase | | Descriptor: | Cutinase | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-04 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3DEA
 
 | | Glomerella cingulata PETFP-cutinase complex | | Descriptor: | 1,1,1-trifluoro-3-[(2-phenylethyl)sulfanyl]propan-2-one, Cutinase | | Authors: | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | Deposit date: | 2008-06-09 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3LKB
 
 | |
3LMV
 
 | | D-Tyr-tRNA(Tyr) Deacylase from plasmodium falciparum in complex with hepes | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-tyrosyl-tRNA(Tyr) deacylase, SULFITE ION | | Authors: | Manickam, Y, Khan, S, Bhatt, T.K, Sharma, A. | | Deposit date: | 2010-02-01 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | Structure of D-tyrosyl-tRNATyr deacylase using home-source Cu Kalpha and moderate-quality iodide-SAD data: structural polymorphism and HEPES-bound enzyme states
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3LOP
 
 | | Crystal structure of substrate-binding periplasmic protein (Pbp) from Ralstonia solanacearum | | Descriptor: | 1,2-ETHANEDIOL, LEUCINE, MAGNESIUM ION, ... | | Authors: | Palani, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-02-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of substrate-binding periplasmic protein (Pbp) from Ralstonia solanacearum
To be Published
|
|
3DEC
 
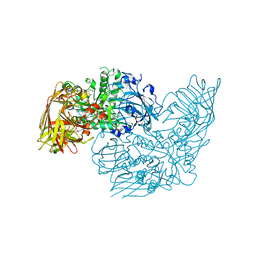 | | Crystal structure of a glycosyl hydrolases family 2 protein from Bacteroides thetaiotaomicron | | Descriptor: | Beta-galactosidase, POTASSIUM ION | | Authors: | Kumaran, D, Bonanno, J, Romero, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-09 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Glycosyl Hydrolases Family 2 protein from Bacteroides thetaiotaomicron.
To be Published
|
|
3LTO
 
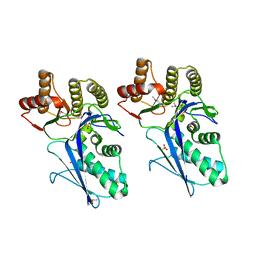 | |
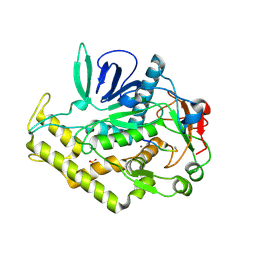
3DDA
 
 | |
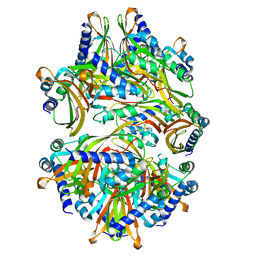
8T17
 
 | |
3DF7
 
 | |
8T18
 
 | |
8T15
 
 | |
8SYG
 
 | |
8T6K
 
 | |
8T6Q
 
 | |
3DDB
 
 | |
3LUA
 
 | |
3DUP
 
 | | Crystal structure of mutt/nudix family hydrolase from rhodospirillum rubrum atcc 11170 | | Descriptor: | GLYCEROL, MutT/nudix family protein, PHOSPHATE ION | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Freeman, J, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-17 | | Release date: | 2008-09-02 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Mutt/Nudix Family Hydrolase from Rhodospirillum Rubrum
To be Published
|
|
3LXT
 
 | |
3DMY
 
 | |
3DH0
 
 | |
3M8N
 
 | |
