5AI9
 
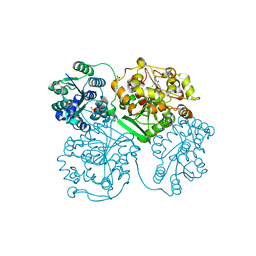 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 2-bromo-4-tert-butyl-6-{[(3-phenylpropyl)amino]methyl}phenol, BIFUNCTIONAL EPOXIDE HYDROLASE 2, GLYCEROL, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-02-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
2MU3
 
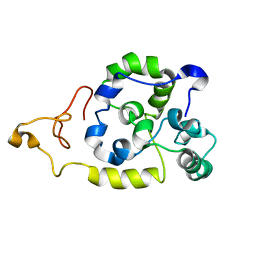 | | Spider wrapping silk fibre architecture arising from its modular soluble protein precursor | | Descriptor: | Aciniform spidroin 1 | | Authors: | Xu, L, Tremblay, M, Meng, Q, Liu, X, Rainey, J.K, Lefevre, T, Sarker, M, Orrell, K.E, Leclerc, J, Pezolet, M, Auger, M. | | Deposit date: | 2014-09-03 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spider wrapping silk fibre architecture arising from its modular soluble protein precursor.
Sci Rep, 5, 2015
|
|
1NHU
 
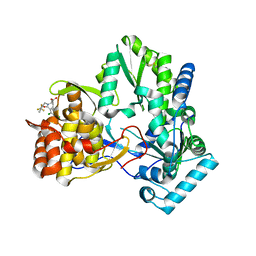 | | Hepatitis C virus RNA polymerase in complex with non-nucleoside analogue inhibitor | | Descriptor: | (2S)-2-[(2,4-DICHLORO-BENZOYL)-(3-TRIFLUOROMETHYL-BENZYL)-AMINO]-3-PHENYL-PROPIONIC ACID, HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Authors: | Wang, M, Ng, K.K.S, Cherney, M.M, Chan, L, Yannopoulos, C.G, Bedard, J, Morin, N, Nguyen-Ba, N, Alaoui-Ismaili, M.H, Bethell, R.C, James, M.N.G. | | Deposit date: | 2002-12-19 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-Nucleoside Analogue Inhibitors Bind to an Allosteric Site on
HCV NS5B Polymerase: Crystal Structures and Mechanism of Inhibition
J.Biol.Chem., 278, 2003
|
|
5AIC
 
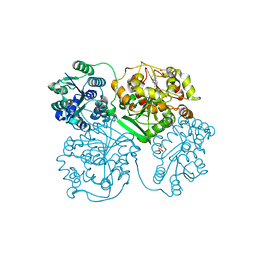 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 7-methyl-2H-1,4-benzothiazin-3(4H)-one, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-02-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5M3A
 
 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-6-(1-methyl-5-phenoxy-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | Authors: | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
5AKZ
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 1-CYCLOHEXYL-3-METHYL-UREA, BIFUNCTIONAL EPOXIDE HYDROLASE 2, GLYCEROL, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
5A40
 
 | | Crystal structure of a dual topology fluoride ion channel. | | Descriptor: | MERCURY (II) ION, MONOBODIES, PUTATIVE FLUORIDE ION TRANSPORTER CRCB | | Authors: | Stockbridge, R.B, Kolmakova-Partensky, L, Shane, T, Koide, A, Koide, S, Miller, C, Newstead, S. | | Deposit date: | 2015-06-04 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal Structures of a Double-Barrelled Fluoride Ion Channel.
Nature, 525, 2015
|
|
1VYH
 
 | | PAF-AH Holoenzyme: Lis1/Alfa2 | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB ALPHA SUBUNIT, PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE IB BETA SUBUNIT | | Authors: | Tarricone, C, Perrina, F, Monzani, S, Massimiliano, L, Knapp, S, Tsai, L.-H, Derewenda, Z.S, Musacchio, A. | | Deposit date: | 2004-04-30 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Coupling Paf Signaling to Dynein Regulation: Structure of Lis1 in Complex with Paf-Acetylhydrolase.
Neuron, 44, 2004
|
|
5AK4
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 3-METHYL-4-PHENYL-1H-PYRAZOLE, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-02 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
7Q94
 
 | | Crystal Structure of Agrobacterium tumefaciens NADQ, DNA complex. | | Descriptor: | DNA binding region (31-MER), NADQ transcription factor | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
1N94
 
 | | Aryl Tetrahydropyridine Inhbitors of Farnesyltransferase: Glycine, Phenylalanine and Histidine Derivates | | Descriptor: | 2-{(5-{[BUTYL-(2-CYCLOHEXYL-ETHYL)-AMINO]-METHYL}-2'-METHYL-BIPHENYL-2-CARBONYL)-AMINO]-4-METHYLSULFANYL-BUTYRIC ACID, ALPHA-HYDROXYFARNESYLPHOSPHONIC ACID, Protein farnesyltransferase alpha subunit, ... | | Authors: | Gwaltney II, S.L, O'Connor, S.J, Nelson, L.T, Sullivan, G.M, Imade, H, Wang, W, Hasvold, L, Li, Q, Cohen, J, Gu, W.Z. | | Deposit date: | 2002-11-22 | | Release date: | 2003-01-07 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Aryl tetrahydropyridine inhibitors of farnesyltransferase: glycine, phenylalanine and histidine derivatives.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1N9H
 
 | | structure of microgravity-grown oxidized myoglobin mutant YQR (ISS6A) | | Descriptor: | HYDROXIDE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miele, A.E, Sciara, G, Federici, L, Vallone, B, Brunori, M. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of the effect of microgravity on protein crystal quality: the case of a myoglobin triple mutant.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5LZK
 
 | | Structure of the domain of unknown function DUF1669 from human FAM83B | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Protein FAM83B | | Authors: | Pinkas, D.M, Bufton, J.C, Williams, E.P, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.575 Å) | | Cite: | Structure of the domain of unknown function DUF1669 from human FAM83B
To Be Published
|
|
7Q91
 
 | | Crystal Structure of Agrobacterium tumefaciens NADQ, native form. | | Descriptor: | NADQ transcription factor, SODIUM ION | | Authors: | Cianci, M, Minazzato, G, Heroux, A, Raffaelli, N, Sorci, L, Gasparrini, M. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Bacterial NadQ (COG4111) is a Nudix-like, ATP-responsive regulator of NAD biosynthesis.
J.Struct.Biol., 214, 2022
|
|
5M6A
 
 | | Crystal structure of cardiotoxic Bence-Jones light chain dimer H9 | | Descriptor: | Bence-Jones light chain, GLYCEROL, PHOSPHATE ION | | Authors: | Oberti, L, Rognoni, P, Bacarizo, J, Bolognesi, M, Ricagno, S. | | Deposit date: | 2016-10-24 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity.
Sci Rep, 7, 2017
|
|
1NEM
 
 | | Saccharide-RNA recognition in the neomycin B / RNA aptamer complex | | Descriptor: | 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranose, 2,6-diamino-2,6-dideoxy-beta-L-idopyranose-(1-3)-beta-D-ribofuranose, 2-DEOXY-D-STREPTAMINE, ... | | Authors: | Jiang, L, Majumdar, A, Hu, W, Jaishree, T.J, Xu, W, Patel, D.J. | | Deposit date: | 1999-03-15 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Saccharide-RNA recognition in a complex formed between neomycin B and an RNA aptamer
Structure Fold.Des., 7, 1999
|
|
5Y9H
 
 | | Crystal structure of SafDAA-dsc complex | | Descriptor: | SafD,Putative outer membrane protein,Putative outer membrane protein,Putative outer membrane protein | | Authors: | Zeng, L.H, Zhang, L, Wang, P.R, Meng, G. | | Deposit date: | 2017-08-25 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of host recognition and biofilm formation by Salmonella Saf pili
Elife, 6, 2017
|
|
1NBA
 
 | | CRYSTAL STRUCTURE ANALYSIS, REFINEMENT AND ENZYMATIC REACTION MECHANISM OF N-CARBAMOYLSARCOSINE AMIDOHYDROLASE FROM ARTHROBACTER SP. AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | N-CARBAMOYLSARCOSINE AMIDOHYDROLASE, SULFATE ION | | Authors: | Romao, M.J, Turk, D, Gomis-Ruth, F.-Z, Huber, R, Schumacher, G, Mollering, H, Russmann, L. | | Deposit date: | 1992-05-18 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis, refinement and enzymatic reaction mechanism of N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
5A8I
 
 | | Crystal structure of the FHA domain of ArnA from Sulfolobus acidocaldarius | | Descriptor: | ARNA, GLYCEROL, SULFATE ION | | Authors: | Hoffmann, L, Anders, K, Reimann, J, Linne, U, Essen, L.-O, Albers, S.-V. | | Deposit date: | 2015-07-16 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phosphorylation-Dependent Interaction between an Archaeal Von Willebrand and Fha Domain Recruits Arna-Arnb Complex to DNA for Repression of Motility
To be Published
|
|
1NEY
 
 | | Triosephosphate Isomerase in Complex with DHAP | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, triosephosphate isomerase | | Authors: | Jogl, G, Rozovsky, S, McDermott, A.E, Tong, L. | | Deposit date: | 2002-12-12 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Optimal alignment for enzymatic proton transfer: Structure of the
Michaelis complex of triosephosphate isomerase at 1.2-A resolution.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5A93
 
 | | 293K Joint X-ray Neutron with Cefotaxime: EXPLORING THE MECHANISM OF BETA-LACTAM RING PROTONATION IN THE CLASS A BETA-LACTAMASE ACYLATION MECHANISM USING NEUTRON AND X-RAY CRYSTALLOGRAPHY | | Descriptor: | BETA-LACTAMASE CTX-M-97, CEFOTAXIME, C3' cleaved, ... | | Authors: | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | NEUTRON DIFFRACTION (1.598 Å), X-RAY DIFFRACTION | | Cite: | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
5MC8
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-D-glucal and alpha-1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
5A1H
 
 | | Crystal structure of human Spindlin3 | | Descriptor: | SPINDLIN-3 | | Authors: | Srikannathasan, V, Gileadi, C, Johansson, C, Shrestha, L, Tallon, R, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2015-04-30 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Spindlin3
To be Published
|
|
1NAZ
 
 | | structure of microgravity-grown oxidized myoglobin mutant YQR (ISS8A) | | Descriptor: | HYDROXIDE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miele, A.E, Federici, L, Sciara, G, Draghi, F, Brunori, M, Vallone, B. | | Deposit date: | 2002-11-30 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Analysis of the effect of microgravity on protein crystal quality: the case of a myoglobin triple mutant.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5YHG
 
 | | The crystal structure of Staphylococcus aureus CntA in complex with staphylopine and zinc | | Descriptor: | (2~{S})-4-[[(2~{R})-3-(1~{H}-imidazol-4-yl)-1-oxidanyl-1-oxidanylidene-propan-2-yl]amino]-2-[[(2~{S})-1-oxidanyl-1-oxidanylidene-propan-2-yl]amino]butanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Ji, Q, Song, L. | | Deposit date: | 2017-09-28 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanistic insights into staphylopine-mediated metal acquisition
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
