7UOC
 
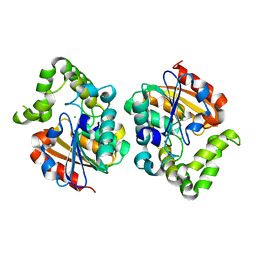 | | Crystal structure of Orobanche minor KAI2d4 | | Descriptor: | CHLORIDE ION, KAI2d4 | | Authors: | Burger, M, Chory, J. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Divergent Clade KAI2 Protein in the Root Parasitic Plant Orobanche minor Is a Highly Sensitive Strigolactone Receptor and Is Involved in the Perception of Sesquiterpene Lactones.
Plant Cell.Physiol., 64, 2023
|
|
6IF5
 
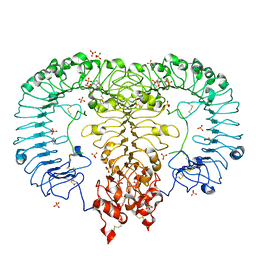 | | Crystal structure of monkey TLR7 in complex with 2',3'-cGMP (Guanosine 2',3'-cyclic phosphate) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GUANOSINE-5'-PHOSPHATE-2',3'-CYCLIC PHOSPHATE, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2018-09-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analyses of Toll-like Receptor 7 Reveal Detailed RNA Sequence Specificity and Recognition Mechanism of Agonistic Ligands.
Cell Rep, 25, 2018
|
|
1V4P
 
 | |
2ZZR
 
 | |
3ABQ
 
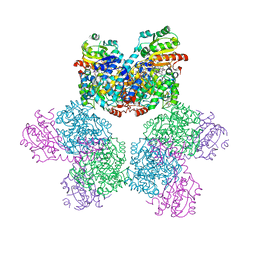 | |
3ABR
 
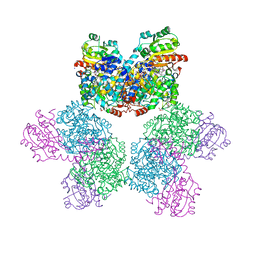 | |
3WO1
 
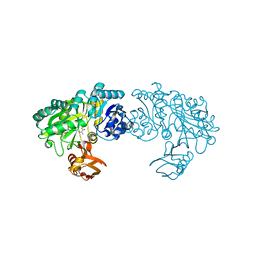 | | Crystal structure of Trp332Ala mutant YwfE, an L-amino acid ligase, with bound ADP-Mg-Ala | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALANINE, Alanine-anticapsin ligase BacD, ... | | Authors: | Tsuda, T, Kojima, S. | | Deposit date: | 2013-12-19 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Single Mutation Alters the Substrate Specificity of l-Amino Acid Ligase
Biochemistry, 53, 2014
|
|
3WF8
 
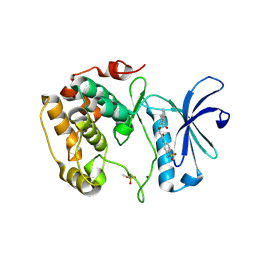 | | Crystal structure of S6K1 kinase domain in complex with a quinoline derivative 2-oxo-2-[(4-sulfamoylphenyl)amino]ethyl 7,8,9,10-tetrahydro-6H-cyclohepta[b]quinoline-11-carboxylate | | Descriptor: | 2-oxo-2-[(4-sulfamoylphenyl)amino]ethyl 7,8,9,10-tetrahydro-6H-cyclohepta[b]quinoline-11-carboxylate, GLYCEROL, Ribosomal protein S6 kinase beta-1, ... | | Authors: | Niwa, H, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Crystal structures of the S6K1 kinase domain in complexes with inhibitors
J.Struct.Funct.Genom., 15, 2014
|
|
3WF9
 
 | | Crystal structure of S6K1 kinase domain in complex with a quinoline derivative 1-oxo-1-[(4-sulfamoylphenyl)amino]propan-2-yl-2-methyl-1,2,3,4-tetrahydroacridine-9-carboxylate | | Descriptor: | (2S)-1-oxo-1-[(4-sulfamoylphenyl)amino]propan-2-yl (2S)-2-methyl-1,2,3,4-tetrahydroacridine-9-carboxylate, GLYCEROL, Ribosomal protein S6 kinase beta-1, ... | | Authors: | Niwa, H, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-06 | | Last modified: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | Crystal structures of the S6K1 kinase domain in complexes with inhibitors
J.Struct.Funct.Genom., 15, 2014
|
|
3X2M
 
 | | X-ray structure of PcCel45A with cellopentaose at 0.64 angstrom resolution. | | Descriptor: | Endoglucanase V-like protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.64 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2O
 
 | | Neutron and X-ray joint refined structure of PcCel45A apo form at 298K. | | Descriptor: | Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Tanaka, I, Niimura, N, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2019-12-18 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2P
 
 | | Neutron and X-ray joint refined structure of PcCel45A with cellopentaose at 298K. | | Descriptor: | Endoglucanase V-like protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Tanaka, I, Niimura, N, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2020-07-29 | | Method: | NEUTRON DIFFRACTION (1.518 Å), X-RAY DIFFRACTION | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2K
 
 | | X-ray structure of PcCel45A D114N with cellopentaose at 95K. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein, ... | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.182 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2I
 
 | | X-ray structure of PcCel45A N92D apo form at 298K. | | Descriptor: | 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2G
 
 | | X-ray structure of PcCel45A N92D apo form at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2H
 
 | | X-ray structure of PcCel45A N92D with cellopentaose at 95K. | | Descriptor: | 3-methylpentane-1,5-diol, Endoglucanase V-like protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2J
 
 | | X-ray structure of PcCel45A D114N apo form at 95K. | | Descriptor: | 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3X2L
 
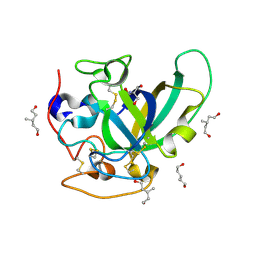 | | X-ray structure of PcCel45A apo form at 95K. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Ohta, K, Tanaka, H, Inaka, K, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
7LUI
 
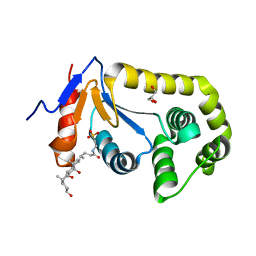 | |
7LSM
 
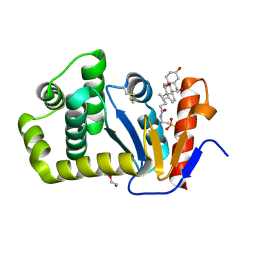 | | Crystal structure of E.coli DsbA in complex with bile salt taurocholate | | Descriptor: | DI(HYDROXYETHYL)ETHER, TAUROCHOLIC ACID, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2021-02-18 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Selective Binding of Small Molecules to Vibrio cholerae DsbA Offers a Starting Point for the Design of Novel Antibacterials.
Chemmedchem, 17, 2022
|
|
7MCA
 
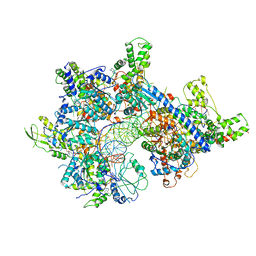 | |
5I8N
 
 | |
4CIU
 
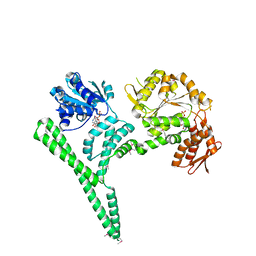 | | Crystal structure of E. coli ClpB | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPERONE PROTEIN CLPB | | Authors: | Kopp, J, Sinning, I, Bukau, B, Kummer, E, Mogk, A. | | Deposit date: | 2013-12-16 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Head-to-Tail Interactions of the Coiled-Coil Domains Regulate Clpb Cooperation with Hsp70 in Protein Disaggregation
Elife, 3, 2014
|
|
376D
 
 | | A ZIPPER-LIKE DNA DUPLEX D(GCGAAAGCT) | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*(CBR)P*GP*AP*AP*AP*GP*CP*T)-3') | | Authors: | Cruse, W.B.T, Shepard, W, Prange, T, delalFortelle, E, Fourme, R. | | Deposit date: | 1998-01-22 | | Release date: | 1999-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A zipper-like duplex in DNA: the crystal structure of d(GCGAAAGCT) at 2.1 A resolution.
Structure, 6, 1998
|
|
3X2N
 
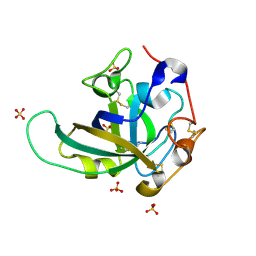 | | Proton relay pathway in inverting cellulase | | Descriptor: | Endoglucanase V-like protein, SULFATE ION | | Authors: | Nakamura, A, Ishida, T, Fushinobu, S, Igarashi, K, Samejima, M. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
