3D3A
 
 | |
3FIJ
 
 | |
3D5L
 
 | |
3F6A
 
 | |
3DEC
 
 | | Crystal structure of a glycosyl hydrolases family 2 protein from Bacteroides thetaiotaomicron | | Descriptor: | Beta-galactosidase, POTASSIUM ION | | Authors: | Kumaran, D, Bonanno, J, Romero, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-09 | | Release date: | 2008-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Glycosyl Hydrolases Family 2 protein from Bacteroides thetaiotaomicron.
To be Published
|
|
3GVX
 
 | |
3DDA
 
 | |
3DF7
 
 | |
3GRC
 
 | |
3IPL
 
 | | CRYSTAL STRUCTURE OF o-succinylbenzoic acid-CoA ligase FROM Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | 2-succinylbenzoate--CoA ligase | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-17 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of o-succinylbenzoic acid-CoA ligase from Staphylococcus aureus
To be Published
|
|
3E9M
 
 | |
3ILV
 
 | |
3IQ0
 
 | |
3EUW
 
 | |
3EDM
 
 | | Crystal structure of a short chain dehydrogenase from Agrobacterium tumefaciens | | Descriptor: | Short chain dehydrogenase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-03 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a short chain dehydrogenase from Agrobacterium tumefaciens
To be Published
|
|
3ILH
 
 | |
3IKH
 
 | |
3ID9
 
 | |
3IE7
 
 | |
8RIJ
 
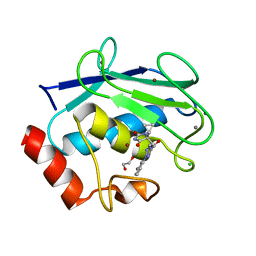 | | Discovery of the first orally bioavailable ADAMTS7 inhibitor BAY-9835 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Schafer, M, Meibom, D, Wasnaire, P, Beyer, K, Broehl, A, Cancho-Grande, Y, Elowe, N, Henninger, K, Johannes, S, Jungmann, N, Krainz, T, Lindner, N, Maassen, S, MacDonald, B, Menshykau, D, Mittendorf, J, Sanchez, G, Stefan, E, Torge, A, Xing, Y, Zubov, D. | | Deposit date: | 2023-12-18 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | BAY-9835: Discovery of the First Orally Bioavailable ADAMTS7 Inhibitor.
J.Med.Chem., 67, 2024
|
|
3C19
 
 | | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein MK0293 | | Authors: | Patskovsky, Y, Romero, R, Bonanno, J.B, Malashkevich, V, Dickey, M, Chang, S, Koss, J, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19.
To be Published
|
|
7YK5
 
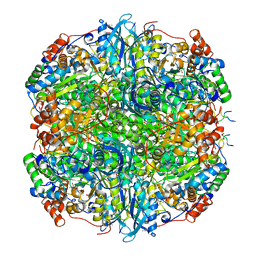 | | Rubisco from Phaeodactylum tricornutum bound to PYCO1(452-592) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, Multifunctional fusion protein, PYCO1 LSU binding motif, ... | | Authors: | Oh, Z.G, Ang, W.S.L, Bhushan, S, Mueller-Cajar, O. | | Deposit date: | 2022-07-21 | | Release date: | 2023-06-21 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | A linker protein from a red-type pyrenoid phase separates with Rubisco via oligomerizing sticker motifs.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8RQ9
 
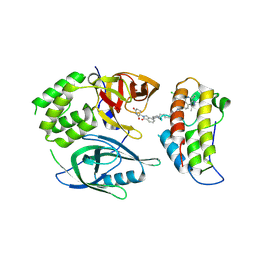 | | Crystal structure of PROTAC CFT-1297 in complex with CRBN-midi and BRD4(BD2) | | Descriptor: | (3~{S})-3-[7-[1-[7-[4-[6-(4-chlorophenyl)-1-methyl-spiro[[1,2,4]triazolo[4,3-a][1,4]benzodiazepine-4,1'-cyclopropane]-8-yl]pyrazol-1-yl]heptyl]piperidin-4-yl]-3-oxidanylidene-1~{H}-isoindol-2-yl]piperidine-2,6-dione, Bromodomain-containing protein 4, Protein cereblon, ... | | Authors: | Darren, D, Ramachandran, S, Kroupova, A, Zollman, D, Ciulli, A. | | Deposit date: | 2024-01-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders.
Nat Commun, 15, 2024
|
|
3C3M
 
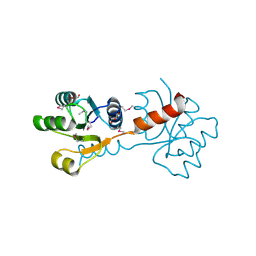 | | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1 | | Descriptor: | GLYCEROL, Response regulator receiver protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Dickey, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1.
To be Published
|
|
3L4A
 
 | |
