7CRK
 
 | | 2ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRX
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (2.63mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRT
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.17mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRS
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.90mJ/mm2) | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BZ9
 
 | | Template lasso peptide C24 mutant I4A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZA
 
 | | Template lasso peptide C24 | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZ8
 
 | | Template lasso peptide C24 mutant V3A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7CU6
 
 | | lasso peptide C24 mutant - A11V2C | | Descriptor: | lasso peptide C24_A11V2C | | Authors: | Ma, M, Liu, X.H. | | Deposit date: | 2020-08-21 | | Release date: | 2021-08-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7EFT
 
 | |
7EMY
 
 | |
7EN1
 
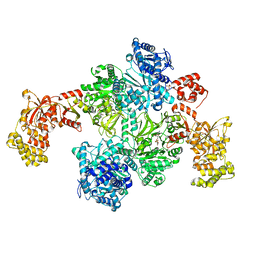 | | Pyochelin synthetase, a dimeric nonribosomal peptide synthetase elongation module-after-condensation | | Descriptor: | (4S)-2-(2-hydroxyphenyl)-4,5-dihydro-1,3-thiazole-4-carboxylic acid, 2-HYDROXYBENZOIC ACID, 4'-PHOSPHOPANTETHEINE, ... | | Authors: | Wang, J.L, Wang, Z.J. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-22 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Catalytic trajectory of a dimeric nonribosomal peptide synthetase subunit with an inserted epimerase domain.
Nat Commun, 13, 2022
|
|
7EN2
 
 | |
3IY9
 
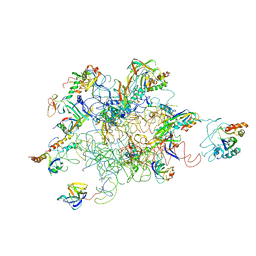 | | Leishmania Tarentolae Mitochondrial Large Ribosomal Subunit Model | | Descriptor: | 39S ribosomal protein L11, mitochondrial, 39S ribosomal protein L16, ... | | Authors: | Sharma, M.R, Booth, T.M, Simpson, L, Maslov, D.A, Agrawal, R.K. | | Deposit date: | 2009-04-20 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Structure of a mitochondrial ribosome with minimal RNA
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7FG9
 
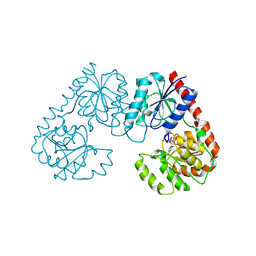 | | Alpha-1,2-glucosyltransferase_UDP_tll1591 | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Su, J.Y. | | Deposit date: | 2021-07-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for glucosylsucrose synthesis by a member of the alpha-1,2-glucosyltransferase family
Acta Biochim.Biophys.Sin., 54, 2022
|
|
7FGA
 
 | | Alpha-1,2-glucosyltransferase_UDP_sucrose_tll1591 | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE, alpha-D-glucopyranose-(1-1)-alpha-D-fructofuranose | | Authors: | Su, J.Y. | | Deposit date: | 2021-07-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for glucosylsucrose synthesis by a member of the alpha-1,2-glucosyltransferase family
Acta Biochim.Biophys.Sin., 54, 2022
|
|
7KPY
 
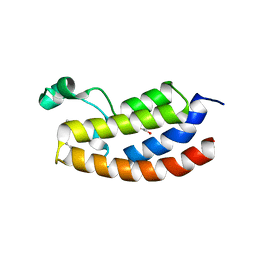 | | Crystal structure of CBP bromodomain liganded with UMB298 (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloranyl-4-methoxy-phenyl)ethyl]-~{N}-cyclohexyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)imidazo[1,2-a]pyridin-3-amine, Histone acetyltransferase | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2020-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of Dimethylisoxazole-Attached Imidazo[1,2- a ]pyridines as Potent and Selective CBP/P300 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7YKS
 
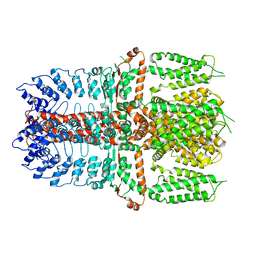 | |
7YKR
 
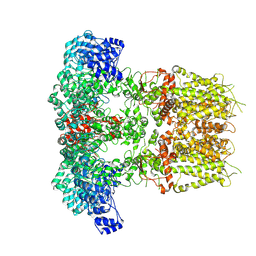 | |
7WVD
 
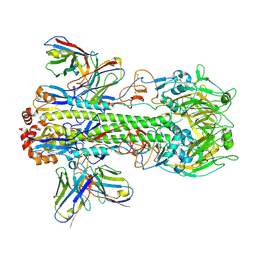 | | Crystal structure of H14 complexed with SIA28 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SIA28, ... | | Authors: | Chen, Y, Qi, J, Gao, G.F. | | Deposit date: | 2022-02-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
7WVI
 
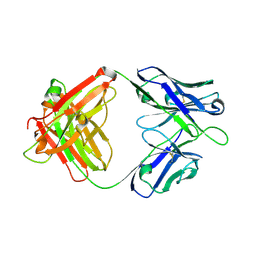 | | Crystal structure of SIA28 | | Descriptor: | Heavy chain of SIA28, Light chain of SIA28 | | Authors: | Chen, Y, Qi, J, Gao, G.F. | | Deposit date: | 2022-02-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
7WVG
 
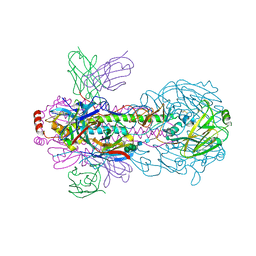 | | Crystal structure of H18 complexed with SIA28 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SIA28, ... | | Authors: | Chen, Y, Qi, J, Gao, G.F. | | Deposit date: | 2022-02-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
3TL1
 
 | | Crystal structure of the Streptomyces coelicolor WhiE ORFVI polyketide aromatase/cyclase | | Descriptor: | 6,7,9-trihydroxy-3-methyl-1H-benzo[g]isochromen-1-one, GLYCEROL, Polyketide cyclase | | Authors: | Lee, M.-Y, Ames, B.D, Zhang, W, Tang, Y, Tsai, S.-C. | | Deposit date: | 2011-08-29 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insight into the Molecular Basis of Aromatic Polyketide Cyclization: Crystal Structure and in Vitro Characterization of WhiE-ORFVI.
Biochemistry, 51, 2012
|
|
7CPY
 
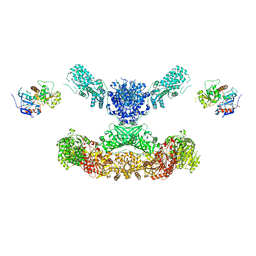 | | Lovastatin nonaketide synthase with LovC | | Descriptor: | Lovastatin nonaketide synthase, enoyl reductase component lovC, polyketide synthase component, ... | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2020-08-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for the biosynthesis of lovastatin.
Nat Commun, 12, 2021
|
|
7BZW
 
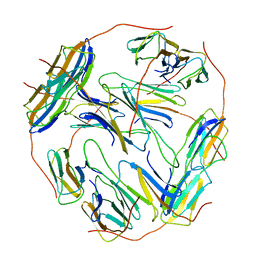 | | Structure of Hsp21 | | Descriptor: | Heat shock protein 21, chloroplastic | | Authors: | Lau, W.C.Y. | | Deposit date: | 2020-04-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of substrate recognition and thermal protection by a small heat shock protein.
Nat Commun, 12, 2021
|
|
7D2G
 
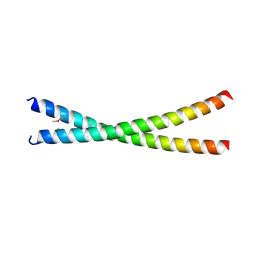 | | Coiled-coil structure of liprin-alpha2_H2delC | | Descriptor: | GLYCEROL, Liprin-alpha-2 | | Authors: | Liang, M, Wei, Z. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oligomerized liprin-alpha promotes phase separation of ELKS for compartmentalization of presynaptic active zone proteins.
Cell Rep, 34, 2021
|
|
