5WQC
 
 | | Crystal structure of human orexin 2 receptor bound to the selective antagonist EMPA determined by the synchrotron light source at SPring-8. | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Hirata, K, Yamashita, K, Tsujimoto, H, Sasanuma, M, Horita, S, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-11-25 | | Release date: | 2017-11-29 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA
Structure, 26, 2018
|
|
7QLL
 
 | |
7QLI
 
 | | Cis structure of rsKiiro at 290 K | | Descriptor: | GLYCEROL, SULFATE ION, rsKiiro | | Authors: | van Thor, J.J, Baxter, J.M. | | Deposit date: | 2021-12-20 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.155 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLK
 
 | |
7QLJ
 
 | |
7QLN
 
 | | rsKiiro pump probe structure by TR-SFX | | Descriptor: | rsKiiro | | Authors: | van Thor, J.J. | | Deposit date: | 2021-12-20 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLO
 
 | | rsKiiro pump dump probe structure by TR-SFX | | Descriptor: | rsKiiro | | Authors: | van Thor, J.J. | | Deposit date: | 2021-12-20 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
7QLM
 
 | |
7URS
 
 | |
7URQ
 
 | |
6PU2
 
 | | Dark, Mutant H275T , 100K, PCM Myxobacterial Phytochrome, P2 | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
6T3L
 
 | | PAS-GAF fragment from Deinococcus radiodurans phytochrome in dark state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Yuan Wahlgren, W, Pandey, S, Schmidt, M, Westenhoff, S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser.
Elife, 9, 2020
|
|
6T3U
 
 | | PAS-GAF fragment from Deinococcus radiodurans phytochrome 1ps after photoexcitation | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Yuan Wahlgren, W, Pandey, S, Schmidt, M, Westenhoff, S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser.
Elife, 9, 2020
|
|
7C13
 
 | | beta1 domain-swapped structure of monothiol cGrx1(C16S) | | Descriptor: | Glutaredoxin, Peptide methionine sulfoxide reductase MsrA | | Authors: | Lee, K, Hwang, K.Y. | | Deposit date: | 2020-05-02 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Monothiol and dithiol glutaredoxin-1 from clostridium oremlandii: identification of domain-swapped structures by NMR, X-ray crystallography and HDX mass spectrometry.
Iucrj, 7, 2020
|
|
7C12
 
 | | beta1 domain-swapped structure of monothiol cGrx1(C16S) | | Descriptor: | Glutaredoxin | | Authors: | Lee, K, Hwang, K.Y. | | Deposit date: | 2020-05-02 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Monothiol and dithiol glutaredoxin-1 from clostridium oremlandii: identification of domain-swapped structures by NMR, X-ray crystallography and HDX mass spectrometry.
Iucrj, 7, 2020
|
|
6PTQ
 
 | | Dark, Room Temperature, PCM Myxobacterial Phytochrome, P2, Wild Type | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, BENZAMIDINE, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
7C10
 
 | | Dithiol cGrx1 | | Descriptor: | Glutaredoxin | | Authors: | Lee, K, Hwang, K.Y. | | Deposit date: | 2020-05-02 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Monothiol and dithiol glutaredoxin-1 from clostridium oremlandii: identification of domain-swapped structures by NMR, X-ray crystallography and HDX mass spectrometry.
Iucrj, 7, 2020
|
|
6PTX
 
 | | Dark, 100K, PCM Myxobacterial Phytochrome, P2, Wild Type, | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Photoreceptor-histidine kinase BphP | | Authors: | Pandey, S, Schmidt, M, Stojkovic, E.A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures.
Struct Dyn., 6, 2019
|
|
8VSG
 
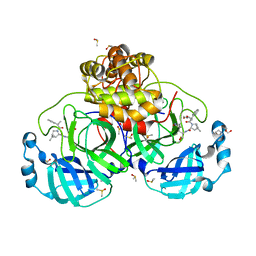 | | SARS-CoV-2 main protease with covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-(1-phenylcyclopropane-1-carbonyl)-3-azabicyclo[3.1.0]hexane-2-carboxamide, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Bell, J.A, Bandera, A.M. | | Deposit date: | 2024-01-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.071 Å) | | Cite: | Exploiting high-energy hydration sites for the discovery of potent peptide aldehyde inhibitors of the SARS-CoV-2 main protease with cellular antiviral activity.
Bioorg.Med.Chem., 103, 2024
|
|
8VQX
 
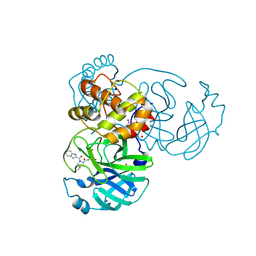 | | Structure of SARS-CoV-2 main protease with potent peptide aldehyde inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dougan, D.R, Lane, W. | | Deposit date: | 2024-01-19 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Exploiting high-energy hydration sites for the discovery of potent peptide aldehyde inhibitors of the SARS-CoV-2 main protease with cellular antiviral activity.
Bioorg.Med.Chem., 103, 2024
|
|
6AZY
 
 | | Crystal structure of Hsp104 R328M/R757M mutant from Calcarisporiella thermophila | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein Hsp104 | | Authors: | Michalska, K, Bigelow, L, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-09-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
4KHB
 
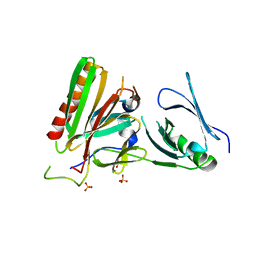 | | Structure of the Spt16D Pob3N heterodimer | | Descriptor: | SULFATE ION, Uncharacterized protein POB3N, Uncharacterized protein SPT16D | | Authors: | Stuwe, T, Zhang, E, Ladurner, A.G. | | Deposit date: | 2013-04-30 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of histone H2A-H2B recognition by the essential chaperone FACT.
Nature, 499, 2013
|
|
6U5C
 
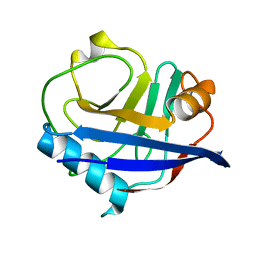 | | RT XFEL structure of CypA solved using MESH injection system | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Wolff, A.M, Thompson, M.C. | | Deposit date: | 2019-08-27 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals
Iucrj, 7, 2020
|
|
6WOR
 
 | |
6TY5
 
 | | Crystal structure of human TLR8 in complex with Compound 11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methyl-7-(7-methyl-2-piperidin-4-yl-indazol-5-yl)furo[3,2-c]pyridin-4-one, Toll-like receptor 8, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Target-Based Identification and Optimization of 5-Indazol-5-yl Pyridones as Toll-like Receptor 7 and 8 Antagonists Using a Biochemical TLR8 Antagonist Competition Assay.
J.Med.Chem., 63, 2020
|
|
