8QHQ
 
 | | Crystal structure of human DNPH1 bound to hmdUMP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
8QHR
 
 | | Crystal structure of the human DNPH1 glycosyl-enzyme intermediate | | Descriptor: | 1',2'-DIDEOXYRIBOFURANOSE-5'-PHOSPHATE, 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, ... | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
8RIL
 
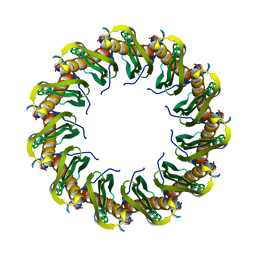 | | Human RAD52 closed ring conformation | | Descriptor: | DNA repair protein RAD52 homolog | | Authors: | Liang, C.C, West, S.C. | | Deposit date: | 2023-12-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of single-stranded DNA annealing by RAD52-RPA complex.
Nature, 629, 2024
|
|
8RK2
 
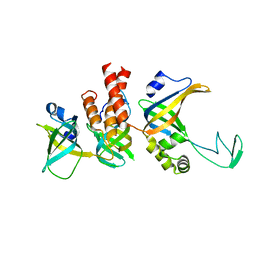 | | Human Replication protein A (RPA; trimeric core) - ssDNA complex | | Descriptor: | Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit, Replication protein A 70 kDa DNA-binding subunit, ... | | Authors: | Liang, C.C, West, S.C. | | Deposit date: | 2023-12-22 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of single-stranded DNA annealing by RAD52-RPA complex.
Nature, 629, 2024
|
|
8RJ3
 
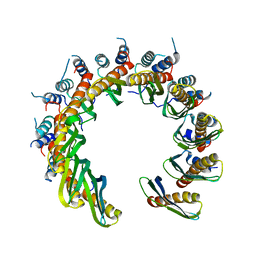 | | Human RAD52 open ring conformation | | Descriptor: | DNA repair protein RAD52 homolog | | Authors: | Liang, C.C, West, S.C. | | Deposit date: | 2023-12-19 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of single-stranded DNA annealing by RAD52-RPA complex.
Nature, 629, 2024
|
|
8RJW
 
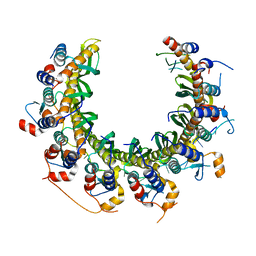 | | Human RAD52 open ring - ssDNA complex | | Descriptor: | DNA repair protein RAD52 homolog, MAGNESIUM ION, ssDNA | | Authors: | Liang, C.C, West, S.C. | | Deposit date: | 2023-12-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Mechanism of single-stranded DNA annealing by RAD52-RPA complex.
Nature, 629, 2024
|
|
5FEY
 
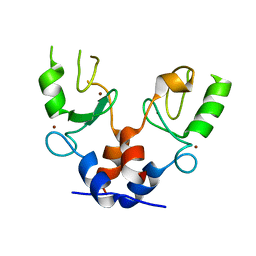 | | TRIM32 RING | | Descriptor: | E3 ubiquitin-protein ligase TRIM32, ZINC ION | | Authors: | Rittinger, K, Esposito, D, Koliopoulos, M.G. | | Deposit date: | 2015-12-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity.
Embo J., 35, 2016
|
|
5FER
 
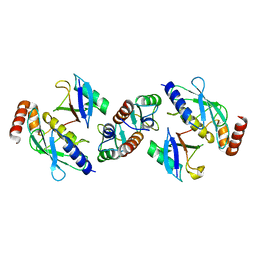 | | Complex of TRIM25 RING with UbcH5-Ub | | Descriptor: | E3 ubiquitin/ISG15 ligase TRIM25, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 D1, ... | | Authors: | Rittinger, K, Koliopoulos, M.G, Esposito, D. | | Deposit date: | 2015-12-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity.
Embo J., 35, 2016
|
|
5HXL
 
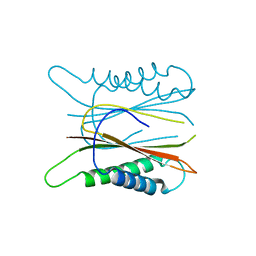 | |
5HYC
 
 | | Structure based function annotation of a hypothetical protein MGG_01005 related to the development of rice blast fungus | | Descriptor: | Cytoplasmic dynein 1 intermediate chain 2, Uncharacterized protein | | Authors: | Liu, J, Li, G, Huang, J, Peng, Y.-l. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure based function-annotation of hypothetical protein MGG_01005 from Magnaporthe oryzae reveals it is the dynein light chain orthologue of dynlt1/3.
Sci Rep, 8, 2018
|
|
4USG
 
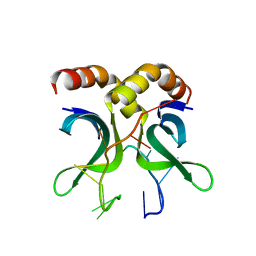 | | Crystal structure of PC4 W89Y mutant complex with DNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP *TP*TP*TP*TP*TP*G)-3', ACTIVATED RNA POLYMERASE II TRANSCRIPTIONAL COACTIVATOR P15 | | Authors: | Zhao, Y, Liu, J. | | Deposit date: | 2014-07-08 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Substitution of Tryptophan 89 with Tyrosine Switches the DNA Binding Mode of Pc4.
Sci.Rep., 5, 2015
|
|
2WQT
 
 | | Dodecahedral assembly of MhpD | | Descriptor: | 2-KETO-4-PENTENOATE HYDRATASE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Montgomery, M.G, Wood, S.P. | | Deposit date: | 2009-08-27 | | Release date: | 2010-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Assembly of a 20Nm Protein Cage by Escherichia Coli 2-Hydroxypentadienoic Acid Hydratase (Mhpd).
J.Mol.Biol., 396, 2010
|
|
4BHM
 
 | | The crystal structure of MoSub1-DNA complex reveals a novel DNA binding mode | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP)-3', MOSUB1 TRANSCRIPTION COFACTOR, SULFATE ION | | Authors: | Huang, J, Liu, H, Zhao, Y, Huang, D, liu, J, Peng, Y. | | Deposit date: | 2013-04-04 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substitution of Tryptophan 89 with Tyrosine Switches the DNA Binding Mode of Pc4.
Sci.Rep., 5, 2015
|
|
4QBL
 
 | | VRR_NUC domain protein | | Descriptor: | MAGNESIUM ION, VRR-NUC | | Authors: | Smerdon, S.J, Pennell, S, Li, J. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | FAN1 activity on asymmetric repair intermediates is mediated by an atypical monomeric virus-type replication-repair nuclease domain.
Cell Rep, 8, 2014
|
|
4QBN
 
 | | VRR_NUC domain | | Descriptor: | Nuclease, SULFATE ION | | Authors: | Smerdon, S.J, Pennell, S, Li, J. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | FAN1 activity on asymmetric repair intermediates is mediated by an atypical monomeric virus-type replication-repair nuclease domain.
Cell Rep, 8, 2014
|
|
4QBO
 
 | | VRR_NUC domain | | Descriptor: | MAGNESIUM ION, Nuclease | | Authors: | Smerdon, S.J, Pennell, S, Li, J. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | FAN1 activity on asymmetric repair intermediates is mediated by an atypical monomeric virus-type replication-repair nuclease domain.
Cell Rep, 8, 2014
|
|
5T3A
 
 | |
