6E0I
 
 | | Crystal structure of Glucokinase in complex with compound 72 | | Descriptor: | 1-{4-[5-({3-[(2-methylpyridin-3-yl)oxy]-5-[(pyridin-2-yl)sulfanyl]pyridin-2-yl}amino)-1,2,4-thiadiazol-3-yl]piperidin-1 -yl}ethan-1-one, DIMETHYL SULFOXIDE, Glucokinase, ... | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
6E0E
 
 | | Crystal structure of Glucokinase in complex with compound 6 | | Descriptor: | 2-({2-[(4-methyl-1,3-thiazol-2-yl)amino]pyridin-3-yl}oxy)benzonitrile, Glucokinase, alpha-D-glucopyranose | | Authors: | Hinklin, R.J, Baer, B.R, Boyd, S.A, Chicarelli, M.D, Condroski, K.R, DeWolf, W.E, Fischer, J, Frank, M, Hingorani, G.P, Lee, P.A, Neitzel, N.A, Pratt, S.A, Singh, A, Sullivan, F.X, Turner, T, Voegtli, W.C, Wallace, E.M, Williams, L, Aicher, T.D. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and preclinical development of AR453588 as an anti-diabetic glucokinase activator.
Bioorg.Med.Chem., 28, 2020
|
|
7XOY
 
 | | Cystathionine beta-synthase of Mycobacterium tuberculosis in the presence of S-adenosylmethionine and serine. | | Descriptor: | Putative cystathionine beta-synthase Rv1077, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Bandyopadhyay, P, Pramanick, I, Biswas, R, Sabarinath, P.S, Sreedharan, S, Singh, S, Rajmani, R, Laxman, S, Dutta, S, Singh, A. | | Deposit date: | 2022-05-01 | | Release date: | 2022-05-25 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | S-Adenosylmethionine-responsive cystathionine beta-synthase modulates sulfur metabolism and redox balance in Mycobacterium tuberculosis.
Sci Adv, 8, 2022
|
|
7XNZ
 
 | | Native cystathionine beta-synthase of Mycobacterium tuberculosis. | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative cystathionine beta-synthase Rv1077 | | Authors: | Bandyopadhyay, P, Pramanick, I, Biswas, R, Sabarinath, P.S, Sreedharan, S, Singh, S, Rajmani, R, Laxman, S, Dutta, S, Singh, A. | | Deposit date: | 2022-04-30 | | Release date: | 2022-05-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | S-Adenosylmethionine-responsive cystathionine beta-synthase modulates sulfur metabolism and redox balance in Mycobacterium tuberculosis.
Sci Adv, 8, 2022
|
|
7XOH
 
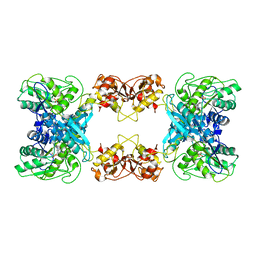 | | Cystathionine beta-synthase of Mycobacterium tuberculosis in the presence of S-adenosylmethionine. | | Descriptor: | Probable cystathionine beta-synthase Rv1077 | | Authors: | Bandyopadhyay, P, Pramanick, I, Biswas, R, Sabarinath, P.S, Sreedharan, S, Singh, S, Rajmani, R, Laxman, S, Dutta, S, Singh, A. | | Deposit date: | 2022-05-01 | | Release date: | 2022-05-25 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | S-Adenosylmethionine-responsive cystathionine beta-synthase modulates sulfur metabolism and redox balance in Mycobacterium tuberculosis.
Sci Adv, 8, 2022
|
|
4JTP
 
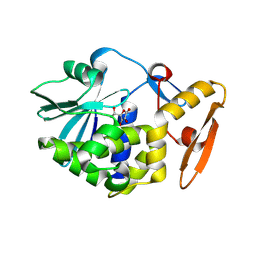 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with Ascorbic acid at 1.85 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBIC ACID, rRNA N-glycosidase | | Authors: | Pandey, S, Bhushan, A, Singh, A, Tyagi, T.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-24 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with Ascorbic acid at 1.85 Angstrom resolution
TO BE PUBLISHED
|
|
4KL4
 
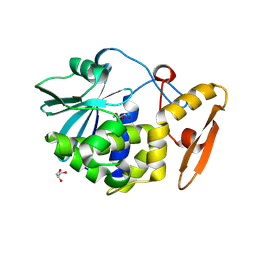 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with Polyethylene glycol at 1.90 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pandey, S, Tyagi, T.K, Singh, A, Bhushan, A, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with Polyethylene glycol at 1.90 Angstrom resolution
To be Published
|
|
3TUS
 
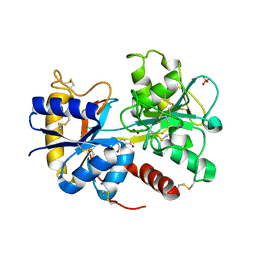 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Meta-hydroxy benzoic acid at 2.5 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-HYDROXYBENZOIC ACID, ... | | Authors: | Shukla, P.K, Gautam, L, Singh, A, Kaushik, S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-09-18 | | Release date: | 2011-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Meta-hydroxy benzoic acid at 2.5 A Resolution
To be Published
|
|
4JTB
 
 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with phosphate ion at 1.71 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, rRNA N-glycosidase | | Authors: | Pandey, S, Tyagi, T.K, Singh, A, Kushwaha, G.S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-23 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with phosphate ion at 1.71 Angstrom resolution
To be published
|
|
4KWN
 
 | | A new stabilizing water structure at the substrate binding site in ribosome inactivating protein from Momordica balsamina at 1.80 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, rRNA N-glycosidase | | Authors: | Yamini, S, Pandey, S, Singh, A, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-24 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A new stabilizing water structure at the substrate binding site in ribosome inactivating protein from Momordica balsamina at 1.80 A resolution
To be Published
|
|
4HSD
 
 | | Crystal structure of a new form of plant lectin from Cicer arietinum at 2.45 Angstrom resolution | | Descriptor: | Lectin, SODIUM ION | | Authors: | Kumar, S, Singh, A, Bhushan, A, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2012-10-30 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a new form of plant lectin from Cicer arietinum at 2.45 Angstrom resolution
to be published
|
|
4RHC
 
 | |
3P2J
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Mycobacterium smegmatis at 2.2 A resolution | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, A, Yadav, R, Sinha, M, Arora, A, Sharma, S, Singh, T.P. | | Deposit date: | 2010-10-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structure of peptidyl-tRNA hydrolase from Mycobacterium smegmatis at 2.2 A resolution
To be Published
|
|
3RT4
 
 | | Structural Basis of Recognition of Pathogen-associated Molecular Patterns and Inhibition of Proinflammatory Cytokines by Camel Peptidoglycan Recognition Protein | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Sharma, P, Dube, D, Singh, A, Mishra, B, Singh, N, Sinha, M, Dey, S, Kaur, P, Mitra, D.K, Sharma, S, Singh, T.P. | | Deposit date: | 2011-05-03 | | Release date: | 2011-06-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Recognition of Pathogen-associated Molecular Patterns and Inhibition of Proinflammatory Cytokines by Camel Peptidoglycan Recognition Protein.
J.Biol.Chem., 286, 2011
|
|
3TTR
 
 | | Crystal structure of C-lobe of bovine lactoferrin complexed with Lidocaine at 2.27 A resolution | | Descriptor: | 2-(diethylamino)-N-(2,6-dimethylphenyl)ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamini, S, Gautam, L, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-09-15 | | Release date: | 2011-10-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of C-lobe of bovine lactoferrin complexed with Lidocaine at 2.27 A resolution
To be Published
|
|
4NZC
 
 | | Crystal structure of Chitinase D from Serratia proteamaculans at 1.45 Angstrom resolution | | Descriptor: | ACETATE ION, GLYCEROL, Glycoside hydrolase family 18 | | Authors: | Madhuprakash, J, Singh, A, Kumar, S, Sinha, M, Kaur, P, Sharma, S, Podile, A.R, Singh, T.P. | | Deposit date: | 2013-12-12 | | Release date: | 2014-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of chitinase D from Serratia proteamaculans reveals the structural basis of its dual action of hydrolysis and transglycosylation
Int J Biochem Mol Biol, 4, 2013
|
|
3RK8
 
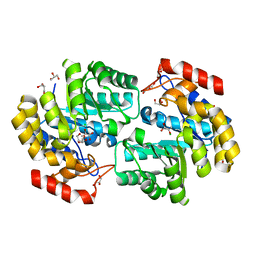 | | Crystal structure of the chloride inhibited dihydrodipicolinate synthase from Acinetobacter baumannii complexed with pyruvate at 1.8 A resolution | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dihydrodipicolinate synthase, ... | | Authors: | Kaushik, S, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-04-17 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the chloride inhibited dihydrodipicolinate synthase from Acinetobacter baumannii complexed with pyruvate at 1.8 A resolution
To be Published
|
|
7CB8
 
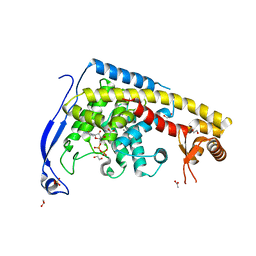 | | Structure of a FIC-domain protein from Mycobacterium marinum in complex with CDP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | Kumar, S, Singh, A, Penmatsa, A, Surolia, A. | | Deposit date: | 2020-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of of a FIC-domain protein from Mycobacterium marinum
To Be Published
|
|
7DSU
 
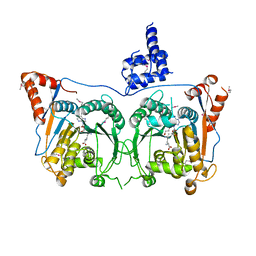 | | Structure of Mod subunit of the Type III restriction-modification enzyme Mbo45V | | Descriptor: | 1,2-ETHANEDIOL, Mbo45V, SINEFUNGIN | | Authors: | Ahmed, I, Chouhan, O.P, Gopinath, A, Morgan, R.D, Bhagat, K, Singh, A, Saikrishnan, K. | | Deposit date: | 2021-01-02 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Mod subunit of the Type III restriction-modification enzyme Mbo45V
To Be Published
|
|
3LP9
 
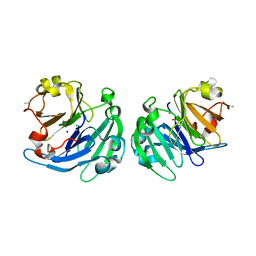 | | Crystal structure of LS24, A Seed Albumin from Lathyrus sativus | | Descriptor: | CALCIUM ION, CHLORIDE ION, LS-24, ... | | Authors: | Gaur, V, Qureshi, I.A, Singh, A, Chanana, V, Salunke, D.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and functional insights of hemopexin fold protein from grass pea
Plant Physiol., 152, 2010
|
|
3ND3
 
 | | Uhelix 16-mer dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*GP*AP*AP*GP*AP*UP*UP*UP*UP*UP*UP*UP*U)-3', POTASSIUM ION, SODIUM ION | | Authors: | Mooers, B.H, Singh, A. | | Deposit date: | 2010-06-07 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The crystal structure of an oligo(U):pre-mRNA duplex from a trypanosome RNA editing substrate.
Rna, 17, 2011
|
|
3ND4
 
 | | Watson-Crick 16-mer dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*GP*AP*AP*GP*AP*UP*CP*UP*UP*CP*UP*CP*U)-3', MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Mooers, B.H, Singh, A. | | Deposit date: | 2010-06-07 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.524 Å) | | Cite: | The crystal structure of an oligo(U):pre-mRNA duplex from a trypanosome RNA editing substrate.
Rna, 17, 2011
|
|
5Y8P
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + 3-Hydroxy propionate (3-HP) | | Descriptor: | (2~{S})-2-methylpentanedioic acid, 3-HYDROXY-PROPANOIC ACID, ACRYLIC ACID, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8I
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate dehydrogenase (MtHIBADH) + (S)-3-hydroxyisobutyrate (S-HIBA) | | Descriptor: | (2S)-2-methyl-3-oxidanyl-propanoic acid, (2~{S})-2-methylpentanedioic acid, GLYCEROL, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
5Y8G
 
 | | Mycobacterium tuberculosis 3-Hydroxyisobutyrate Dehydrogenase (MtHIBADH) | | Descriptor: | (2~{S})-2-methylpentanedioic acid, ACRYLIC ACID, GLYCEROL, ... | | Authors: | Srikalaivani, R, Singh, A, Surolia, A, Vijayan, M. | | Deposit date: | 2017-08-21 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure, interactions and action ofMycobacterium tuberculosis3-hydroxyisobutyric acid dehydrogenase.
Biochem. J., 475, 2018
|
|
