6QHD
 
 | | Lysine acetylated and tyrosine phosphorylated STAT3 in a complex with DNA | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*TP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*CP*T)-3'), Signal transducer and activator of transcription 3 | | Authors: | Arbely, E, Belo, Y, Shahar, A, Zarivach, R. | | Deposit date: | 2019-01-16 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Unexpected implications of STAT3 acetylation revealed by genetic encoding of acetyl-lysine.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
7O85
 
 | | Anthrax toxin prepore in complex with the neutralizing Fab cAb29 | | Descriptor: | CALCIUM ION, Fab, Protective antigen PA-63 | | Authors: | Hoelzgen, F, Zalk, R, Alcalay, R, Cohen-Schwartz, S, Garau, G, Shahar, A, Mazor, O, Frank, G.A. | | Deposit date: | 2021-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Neutralization of the anthrax toxin by antibody-mediated stapling of its membrane-penetrating loop.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8QU9
 
 | | Structure of the NCOA4 (Nuclear Receptor Coactivator 4)-FTH1 (H-Ferritin) complex | | Descriptor: | FE (III) ION, Ferritin heavy chain, NCOA4 (Nuclear Receptor Coactivator 4) | | Authors: | Hoelzgen, F, Klukin, E, Zalk, R, Shahar, A, Cohen-Schwartz, S, Frank, G.A. | | Deposit date: | 2023-10-15 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for the intracellular regulation of ferritin degradation.
Nat Commun, 15, 2024
|
|
1XHL
 
 | | Crystal Structure of putative Tropinone Reductase-II from Caenorhabditis Elegans with Cofactor and Substrate | | Descriptor: | 8-METHYL-8-AZABICYCLO[3,2,1]OCTAN-3-ONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase family member (5L265), ... | | Authors: | Schormann, N, Karpova, E, Zhou, J, Zhang, Y, Symersky, J, Bunzel, R, Huang, W.-Y, Arabshahi, A, Qiu, S, Luan, C.-H, Gray, R, Carson, M, Tsao, J, Luo, M, Johnson, D, Lu, S, Lin, G, Luo, D, Cao, Z, Li, S, McKInstry, A, Shang, Q, Chen, Y.-J, Bray, T, Nagy, L, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of putative Tropinone Reductase-II from Caenorhabditis Elegans with Cofactor and Substrate
To be Published
|
|
1YIS
 
 | | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase | | Descriptor: | SULFATE ION, adenylosuccinate lyase | | Authors: | Symersky, J, Schormann, N, Lu, S, Zhang, Y, Karpova, E, Qiu, S, Huang, W, Cao, Z, Zhou, J, Luo, M, Arabshahi, A, McKinstry, A, Luan, C.-H, Luo, D, Johnson, D, An, J, Tsao, J, Delucas, L, Shang, Q, Gray, R, Li, S, Bray, T, Chen, Y.-J, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase
To be Published
|
|
1XKQ
 
 | | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, short-chain reductase family member (5D234) | | Authors: | Schormann, N, Zhou, J, Karpova, E, Zhang, Y, Symersky, J, Bunzel, B, Huang, W.-Y, Arabshahi, A, Qiu, S, Luan, C.-H, Gray, R, Carson, M, Tsao, J, Luo, M, Johnson, D, Lu, S, Lin, G, Luo, D, Cao, Z, Li, S, McKinstry, A, Shang, Q, Chen, Y.-J, Bray, T, Nagy, L, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-29 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor
To be Published
|
|
6S3G
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant A187C/F291C | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Bash, Y, Rush, I, Shahar, A, Pazy, Y, Fishman, A. | | Deposit date: | 2019-06-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bridges to Stability: Engineering Disulfide Bonds Towards Enhanced Lipase Biodiesel Synthesis
Chemcatchem, 2019
|
|
6S3J
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant E134C/F149C | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Bash, Y, Rush, I, Shahar, A, Pazy, Y, Fishman, A. | | Deposit date: | 2019-06-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bridges to Stability: Engineering Disulfide Bonds Towards Enhanced Lipase Biodiesel Synthesis
Chemcatchem, 2019
|
|
6S3V
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant E251C/G332C | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Bash, Y, Rush, I, Shahar, A, Pazy, Y, Fishman, A. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bridges to Stability: Engineering Disulfide Bonds Towards Enhanced Lipase Biodiesel Synthesis
Chemcatchem, 2019
|
|
8DV3
 
 | | Crystal structure of human CD1b presenting Phosphatidylinositol C34:1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Farquhar, R, Rossjohn, J, Shahine, A. | | Deposit date: | 2022-07-28 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | alpha beta T-cell receptor recognition of self-phosphatidylinositol presented by CD1b.
J.Biol.Chem., 299, 2023
|
|
8DV4
 
 | | Crystal structure of the BC8B TCR-CD1b-PI complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Farquhar, R, Rossjohn, J, Shahine, A. | | Deposit date: | 2022-07-28 | | Release date: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | alpha beta T-cell receptor recognition of self-phosphatidylinositol presented by CD1b.
J.Biol.Chem., 299, 2023
|
|
8GLI
 
 | | Crystal Structure of Human CD1b in Complex with Mycobacterial C85-GMM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-[(2R,3R)-3-hydroxy-20-{(1R,2S)-2-[(17R,18R)-17-methoxy-18-methyldotriacontyl]cyclopropyl}-2-pentadecylicosanoyl]-alpha-L-galactopyranose, Beta-2-microglobulin, ... | | Authors: | Balaji, G, Rossjohn, J, Shahine, A. | | Deposit date: | 2023-03-22 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CD1 lipidomes reveal lipid-binding motifs and size-based antigen-display mechanisms.
Cell, 186, 2023
|
|
8GLH
 
 | | Crystal Structure of Human CD1b in Complex with Endogenous PC C40:5 | | Descriptor: | (11E)-hexadec-11-enoic acid, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Farquhar, R, Rossjohn, J, Shahine, A. | | Deposit date: | 2023-03-22 | | Release date: | 2023-09-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | CD1 lipidomes reveal lipid-binding motifs and size-based antigen-display mechanisms.
Cell, 186, 2023
|
|
7MX4
 
 | | CD1c with antigen analogue 1 | | Descriptor: | (2S)-2,3-dihydroxypropyl hexadecanoate, (4S,8R,12S,16S,20S)-4,8,12,16,20-pentamethylheptacosyl (1R,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl hydrogen (R)-phosphate, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cao, T.P, Shahine, A, Rossjohn, J. | | Deposit date: | 2021-05-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Rational design of a hydrolysis-resistant mycobacterial phosphoglycolipid antigen presented by CD1c to T cells.
J.Biol.Chem., 297, 2021
|
|
7MXF
 
 | | CD1c with antigen analogue 2 | | Descriptor: | (2S)-2,3-dihydroxypropyl hexadecanoate, 2,6-anhydro-1-deoxy-1-[(S)-hydroxy{[(4R,8S,12R,16R,20S)-4,8,12,16,20-pentamethylheptacosyl]oxy}phosphoryl]-D-glycero-D-galacto-heptitol, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Cao, T.P, Shahine, A, Rossjohn, J. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of a hydrolysis-resistant mycobacterial phosphoglycolipid antigen presented by CD1c to T cells.
J.Biol.Chem., 297, 2021
|
|
7MXH
 
 | | CD1c with antigen analogue 3 | | Descriptor: | (2S)-2,3-dihydroxypropyl hexadecanoate, 2,6-anhydro-1-deoxy-1,1-difluoro-1-[(R)-hydroxy{[(4S,8S,12S,16S,20S)-4,8,12,16,20-pentamethylheptacosyl]oxy}phosphoryl]-D-glycero-D-galacto-heptitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cao, T.P, Shahine, A, Rossjohn, J. | | Deposit date: | 2021-05-19 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Rational design of a hydrolysis-resistant mycobacterial phosphoglycolipid antigen presented by CD1c to T cells.
J.Biol.Chem., 297, 2021
|
|
7ZVD
 
 | | K89 acetylated glucose-6-phosphate dehydrogenase (G6PD) in a complex with structural NADP+ | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wu, F, Muskat, N.H, Nudelman, H, Shahar, A, Arbely, E. | | Deposit date: | 2022-05-15 | | Release date: | 2023-08-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Acetylation-dependent coupling between G6PD activity and apoptotic signaling.
Nat Commun, 14, 2023
|
|
7ZVE
 
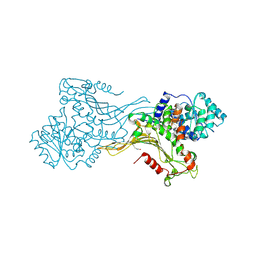 | | K403 acetylated glucose-6-phosphate dehydrogenase (G6PD) | | Descriptor: | COPPER (II) ION, GLYCEROL, Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wu, F, Muskat, N.H, Shahar, A, Arbely, E. | | Deposit date: | 2022-05-15 | | Release date: | 2023-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Acetylation-dependent coupling between G6PD activity and apoptotic signaling.
Nat Commun, 14, 2023
|
|
5JAE
 
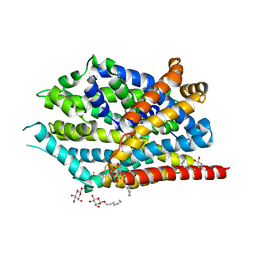 | | LeuT in the outward-oriented, Na+-free return state, P21 form at pH 6.5 | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
7QTV
 
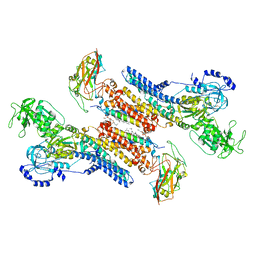 | | Beryllium fluoride form of the Na+,K+-ATPase (E2-BeFx) | | Descriptor: | 1-O-decanoyl-beta-D-tagatofuranosyl beta-D-allopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shahsavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | Deposit date: | 2022-01-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
5JAG
 
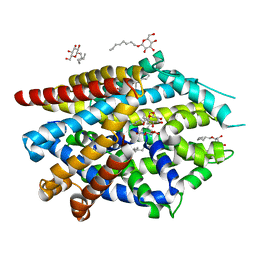 | | LeuT T354H mutant in the outward-oriented, Na+-free Return State | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
5JAF
 
 | | LeuT Na+-free Return State, C2 form at pH 5 | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.021 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
8PVQ
 
 | | Crystal Structure of Human PTX3 C-terminal domain | | Descriptor: | Pentraxin-related protein PTX3 | | Authors: | Levy, C.W. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Pentraxin 3 mediated crosslinking of heavy chain hyaluronan complexes: new insights from structural and biophysical studies
To Be Published
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
6D64
 
 | | Crystal Structure of Human CD1b in Complex with POPC | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shahine, A.E, Rossjohn, J. | | Deposit date: | 2018-04-20 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A T-cell receptor escape channel allows broad T-cell response to CD1b and membrane phospholipids.
Nat Commun, 10, 2019
|
|
