4U3Z
 
 | | APO MAP4K4 T181E Phosphomimetic Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase kinase kinase kinase 4, SODIUM ION | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
4U40
 
 | |
4U42
 
 | | MAP4K4 T181E Mutant Bound to inhibitor compound 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(3S)-3-(4-methyl-1H-pyrazol-3-yl)piperidin-1-yl]pyrido[3,2-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
4U3Y
 
 | |
4U41
 
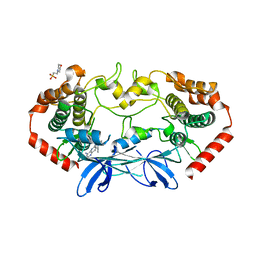 | | MAP4K4 Bound to inhibitor compound 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(3S)-3-(4-methyl-1H-pyrazol-3-yl)piperidin-1-yl]pyrido[3,2-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
3BAT
 
 | |
3BAS
 
 | |
1SCM
 
 | |
5NP3
 
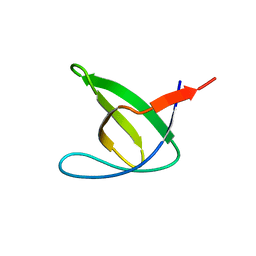 | | Abl2 SH3 | | Descriptor: | Abelson tyrosine-protein kinase 2 | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
5NP5
 
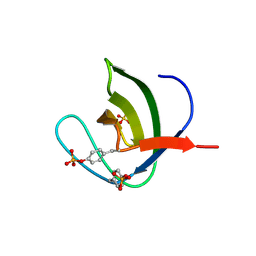 | | Abl2 SH3 pTyr116/161 | | Descriptor: | Abelson tyrosine-protein kinase 2, SULFATE ION | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
8EQ6
 
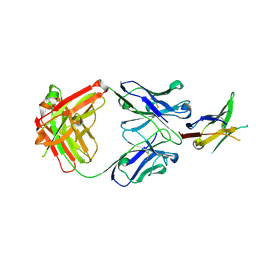 | | PD1 signaling receptor bound to FAB Complex | | Descriptor: | Antibody FAB heavy chain, Antibody FAB light chain, Programmed cell death protein 1 | | Authors: | Bjorkelid, C, Paluch, C, Robertson, N.J. | | Deposit date: | 2022-10-07 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Antibody agonists trigger immune receptor signaling through local exclusion of receptor-type protein tyrosine phosphatases.
Immunity, 57, 2024
|
|
5NP2
 
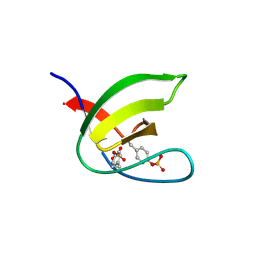 | | Abl1 SH3 pTyr89/134 | | Descriptor: | Tyrosine-protein kinase ABL1 | | Authors: | Mero, B, Radnai, L, Gogl, G, Leveles, I, Buday, L. | | Deposit date: | 2017-04-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the tyrosine phosphorylation-mediated inhibition of SH3 domain-ligand interactions.
J.Biol.Chem., 294, 2019
|
|
2FXO
 
 | | Structure of the human beta-myosin S2 fragment | | Descriptor: | Myosin heavy chain, cardiac muscle beta isoform | | Authors: | Blankenfeldt, W, Thoma, N.H, Wray, J.S, Gautel, M, Schlichting, I. | | Deposit date: | 2006-02-06 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human cardiac {beta}-myosin II S2-{Delta} provide insight into the functional role of the S2 subfragment
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FXM
 
 | | Structure of the human beta-myosin S2 fragment | | Descriptor: | MERCURY (II) ION, Myosin heavy chain, cardiac muscle beta isoform | | Authors: | Blankenfeldt, W, Thoma, N.H, Wray, J.S, Gautel, M, Schlichting, I. | | Deposit date: | 2006-02-06 | | Release date: | 2006-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of human cardiac {beta}-myosin II S2-{Delta} provide insight into the functional role of the S2 subfragment
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7QQL
 
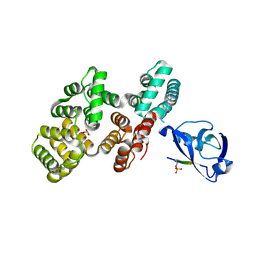 | | The PDZ domain of SNTG2 complexed with the phosphorylated PDZ-binding motif of RSK1 | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-2-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2022-01-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QQN
 
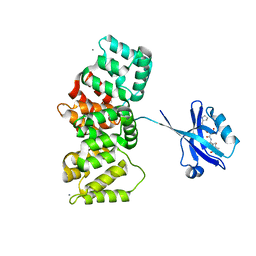 | | The PDZ domain of SNTG1 complexed with the acetylated PDZ-binding motif of TRPV3 | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-1-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2022-01-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QQM
 
 | | The PDZ domain of LRRC7 fused with ANXA2 | | Descriptor: | CALCIUM ION, GLYCEROL, Leucine-rich repeat-containing protein 7,Annexin A2 | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2022-01-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7OLX
 
 | | MerTK kinase domain with type 1.5 inhibitor containing a tri-methyl pyrazole group | | Descriptor: | CHLORIDE ION, Tyrosine-protein kinase Mer, ~{N}-[[3-[4-[(dimethylamino)methyl]phenyl]imidazo[1,2-a]pyridin-6-yl]methyl]-~{N}-methyl-5-[3-methyl-5-(1,3,5-trimethylpyrazol-4-yl)pyridin-2-yl]-1,3,4-oxadiazol-2-amine | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
7OLS
 
 | | MerTK kinase domain with type 1.5 inhibitor containing a di-methyl pyrazole group | | Descriptor: | 5-[4-(1,5-dimethylpyrazol-4-yl)-2-methyl-phenyl]-~{N}-(imidazo[1,2-a]pyridin-6-ylmethyl)-~{N}-methyl-1,3,4-oxadiazol-2-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
7OLV
 
 | | MerTK kinase domain with type 1.5 inhibitor containing a di-methyl, cyano pyrazole group | | Descriptor: | 4-[4-[5-[imidazo[1,2-a]pyridin-6-ylmethyl(methyl)amino]-1,3,4-oxadiazol-2-yl]-3-methyl-phenyl]-2,5-dimethyl-pyrazole-3-carbonitrile, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
5K4L
 
 | |
7PC5
 
 | | The third PDZ domain of PDZD7 complexed with the PDZ-binding motif of EXOC4 | | Descriptor: | CALCIUM ION, Exocyst complex component 4, GLYCEROL, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PCB
 
 | | The PDZ domain of SNX27 fused with ANXA2 | | Descriptor: | CALCIUM ION, GLYCEROL, Sorting nexin-27,Annexin A2 | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PC8
 
 | | The PDZ domain of SNTG1 complexed with the phosphomimetic mutant PDZ-binding motif of RSK1 | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-1-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PC7
 
 | | The PDZ domain of SNTG1 complexed with the acetylated PDZ-binding motif of PTEN | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-1-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
