1KFN
 
 | |
1KFM
 
 | |
9B4W
 
 | | 4F-Trp labeled Oscillatoria Agardhii Agglutinin (OAA) | | Descriptor: | Lectin | | Authors: | Runge, B.R, Zadorozhnyi, R.R, Quinn, C.M, Russell, R.W, Lu, M, Antolinez, S, Struppe, J, Schwieters, C.D, Byeon, I.L, Hadden-Perilla, J, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2024-03-21 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-20 | | Method: | SOLID-STATE NMR | | Cite: | Integrating 19 F Distance Restraints for Accurate Protein Structure Determination by Magic Angle Spinning NMR Spectroscopy.
J.Am.Chem.Soc., 146, 2024
|
|
9B4V
 
 | | 4F-Trp labeled Oscillatoria Agardhii Agglutinin (OAA) | | Descriptor: | Lectin | | Authors: | Runge, B.R, Zadorozhnyi, R.R, Quinn, C.M, Russell, R.W, Lu, M, Antolinez, S, Struppe, J, Schwieters, C.D, Byeon, I.L, Hadden-Perilla, J, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2024-03-21 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-20 | | Method: | SOLID-STATE NMR | | Cite: | Integrating 19 F Distance Restraints for Accurate Protein Structure Determination by Magic Angle Spinning NMR Spectroscopy.
J.Am.Chem.Soc., 146, 2024
|
|
3N27
 
 | |
3U91
 
 | |
1F23
 
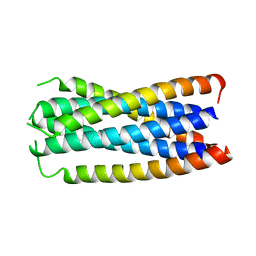 | | CONTRIBUTION OF A BURIED HYDROGEN BOND TO HIV-1 ENVELOPE GLYCOPROTEIN STRUCTURE AND FUNCTION | | Descriptor: | TRANSMEMBRANE GLYCOPROTEIN | | Authors: | Liu, J, Shu, W, Fagan, M, Nunberg, J.H, Lu, M. | | Deposit date: | 2000-05-23 | | Release date: | 2001-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of the HIV gp41 core containing an Ile573 to Thr substitution: implications for membrane fusion.
Biochemistry, 40, 2001
|
|
3BT6
 
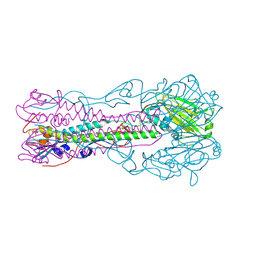 | | Crystal Structure of Influenza B Virus Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Q, Cheng, F, Lu, M, Tian, X, Ma, J. | | Deposit date: | 2007-12-27 | | Release date: | 2008-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of unliganded influenza B virus hemagglutinin.
J.Virol., 82, 2008
|
|
7DHY
 
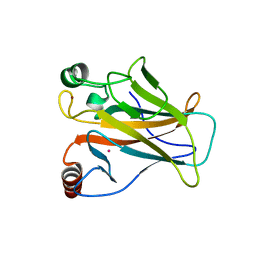 | | Arsenic-bound p53 DNA-binding domain mutant G245S | | Descriptor: | ARSENIC, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Chen, S, Lu, M. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Arsenic Trioxide Rescues Structural p53 Mutations through a Cryptic Allosteric Site.
Cancer Cell, 39, 2021
|
|
7DHZ
 
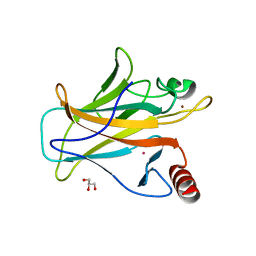 | | Arsenic-bound p53 DNA-binding domain mutant R249S | | Descriptor: | ARSENIC, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Chen, S, Lu, M. | | Deposit date: | 2020-11-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Arsenic Trioxide Rescues Structural p53 Mutations through a Cryptic Allosteric Site.
Cancer Cell, 39, 2021
|
|
3UIA
 
 | |
4DZU
 
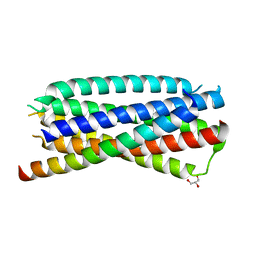 | | Complex of 3-alpha bound to gp41-5 | | Descriptor: | 3-alpha, GLYCEROL, gp41-5 | | Authors: | Johnson, L.M, Mortenson, D.E, Yun, H.G, Horne, W.S, Ketas, T.J, Lu, M, Moore, J.P, Gellman, S.H. | | Deposit date: | 2012-03-01 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancement of alpha-helix mimicry by an alpha / beta-peptide foldamer via incorporation of a dense ionic side-chain array.
J.Am.Chem.Soc., 134, 2012
|
|
4DZV
 
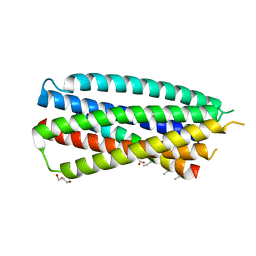 | | Complex of 4-alpha/beta bound to gp41-5 | | Descriptor: | 4-alpha/beta, GLYCEROL, gp41-5 | | Authors: | Johnson, L.M, Mortenson, D.E, Yun, H.G, Horne, W.S, Ketas, T.J, Lu, M, Moore, J.P, Gellman, S.H. | | Deposit date: | 2012-03-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancement of alpha-helix mimicry by an alpha / beta-peptide foldamer via incorporation of a dense ionic side-chain array.
J.Am.Chem.Soc., 134, 2012
|
|
3HCS
 
 | | Crystal structure of the N-terminal domain of TRAF6 | | Descriptor: | TNF receptor-associated factor 6, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HCT
 
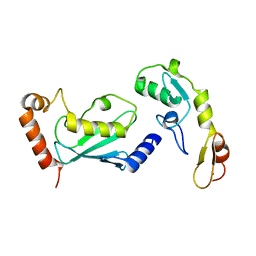 | | Crystal structure of TRAF6 in complex with Ubc13 in the P1 space group | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin-conjugating enzyme E2 N, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
8XCE
 
 | | cocrystal structure of p53 in complex of B23 | | Descriptor: | 2-[(2~{Z})-2-(3-ethynylphenyl)imino-4-oxidanylidene-1,3-thiazolidin-5-ylidene]ethanoic acid, Cellular tumor antigen p53, ZINC ION | | Authors: | Wang, Z, Shujun, X, Yigang, T, Derun, Z, Jiaqi, W, Kai, K, Lu, M. | | Deposit date: | 2023-12-08 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | cocrystal structure of p53 in complex of B23
To Be Published
|
|
8XCB
 
 | | Cocrystal structure of p53 in complex of B20 | | Descriptor: | 2-[2,4-bis(oxidanylidene)-1,3-thiazolidin-5-ylidene]ethanoic acid, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Wang, Z, Shujun, X, Yigang, T, Derun, Z, Jiaqi, W, Kai, K, Lu, M. | | Deposit date: | 2023-12-08 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Cocrystal structure of p53 in complex of B20
To Be Published
|
|
3G9R
 
 | |
2IPZ
 
 | | A Parallel Coiled-Coil Tetramer with Offset Helices | | Descriptor: | GLYCEROL, General control protein GCN4, ISOPROPYL ALCOHOL | | Authors: | Liu, J, Lu, M. | | Deposit date: | 2006-10-12 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Parallel Coiled-Coil Tetramer with Offset Helices.
Biochemistry, 45, 2006
|
|
3HCU
 
 | | Crystal structure of TRAF6 in complex with Ubc13 in the C2 space group | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin-conjugating enzyme E2 N, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
6U2S
 
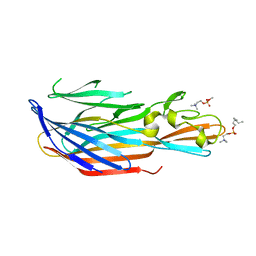 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Bi-component leukocidin LukED subunit D, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U33
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Bi-component leukocidin LukED subunit D, NICKEL (II) ION | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U3T
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U4P
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-26 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
2B1F
 
 | | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat | | Descriptor: | General control protein GCN4 | | Authors: | Deng, Y, Liu, J, Zheng, Q, Eliezer, D, Kallenbach, N.R, Lu, M. | | Deposit date: | 2005-09-15 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antiparallel four-stranded coiled coil specified by a 3-3-1 hydrophobic heptad repeat.
Structure, 14, 2006
|
|
