2ZXD
 
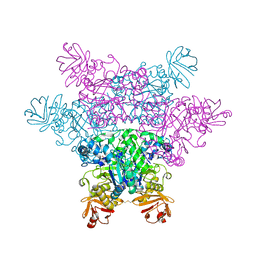 | | alpha-L-fucosidase complexed with inhibitor, iso-6FNJ | | Descriptor: | (2S,3R,4S,5R)-2-(1-methylethyl)piperidine-3,4,5-triol, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX7
 
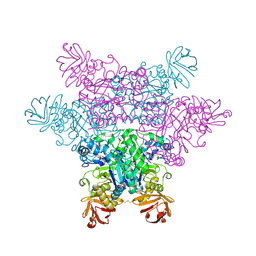 | | alpha-L-fucosidase complexed with inhibitor, F10-2C | | Descriptor: | Alpha-L-fucosidase, putative, N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}-1H-indole-2-carboxamide | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX8
 
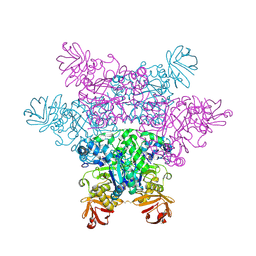 | | alpha-L-fucosidase complexed with inhibitor, F10-2C-O | | Descriptor: | Alpha-L-fucosidase, putative, N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}-1-benzofuran-2-carboxamide | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX6
 
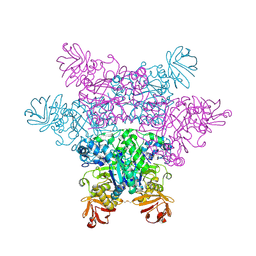 | | alpha-L-fucosidase complexed with inhibitor, F10-1C | | Descriptor: | 2-(1H-indol-3-yl)-N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}acetamide, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX9
 
 | | alpha-L-fucosidase complexed with inhibitor, B4 | | Descriptor: | (2S)-2-cyclopentyl-2-phenyl-N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}ethanamide, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZE3
 
 | |
7C8T
 
 | | Complex Structure of SARS-CoV-2 3CL Protease with TG-0205221 | | Descriptor: | 3C-like proteinase, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Lee, C.C, Wang, A.H.J, Kuo, C.J, Liang, P.H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Complex Structures and Cellular Activities of the Potent SARS-CoV-2 3CLpro Inhibitors Guiding Drug Discovery Against COVID-19
To Be Published
|
|
7C8R
 
 | | Complex Structure of SARS-CoV-2 3CL Protease with TG-0203770 | | Descriptor: | 3C-like proteinase, ethyl (4R)-4-[[(2S)-4-methyl-2-[[(2S,3R)-3-[(2-methylpropan-2-yl)oxy]-2-(phenylmethoxycarbonylamino)butanoyl]amino]pentanoyl]amino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Lee, C.C, Wang, A.H.J, Kuo, C.J, Liang, P.H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex Structures and Cellular Activities of the Potent SARS-CoV-2 3CLpro Inhibitors Guiding Drug Discovery Against COVID-19
To Be Published
|
|
2F4B
 
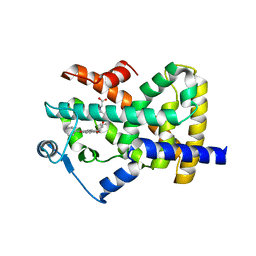 | | Crystal structure of the ligand binding domain of human PPAR-gamma in complex with an agonist | | Descriptor: | (5-{3-[(6-BENZOYL-1-PROPYL-2-NAPHTHYL)OXY]PROPOXY}-1H-INDOL-1-YL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Lu, I.L, Peng, Y.H, Mahindroo, N, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2005-11-23 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Indol-1-yl Acetic Acids as Peroxisome Proliferator-Activated Receptor Agonists: Design, Synthesis, Structural Biology, and Molecular Docking Studies
J.Med.Chem., 49, 2006
|
|
7QQR
 
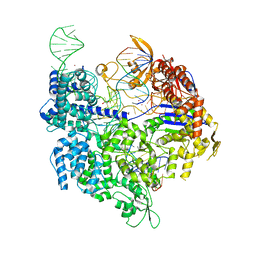 | | SpCas9 bound to AAVS1 off-target5 DNA substrate | | Descriptor: | 1,2-ETHANEDIOL, AAVS1 off-target5 non-target strand, AAVS1 off-target5 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQV
 
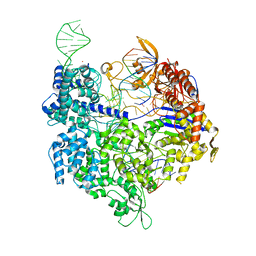 | | SpCas9 bound to FANCF off-target3 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target3 non-target strand, FANCF off-target3 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QR5
 
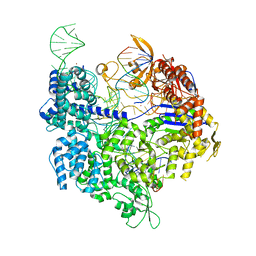 | | SpCas9 bound to FANCF off-target6 DNA substrate | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target6 non-target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQO
 
 | | SpCas9 bound to AAVS1 off-target1 DNA substrate | | Descriptor: | AAVS1 off-target1 non-target strand, AAVS1 off-target1 target strand, AAVS1 sgRNA, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQQ
 
 | | SpCas9 bound to AAVS1 off-target4 DNA substrate | | Descriptor: | 1,2-ETHANEDIOL, AAVS1 off-target4 non-target strand, AAVS1 off-target4 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQU
 
 | | SpCas9 bound to FANCF off-target2 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target2 non-target strand, FANCF off-target2 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQT
 
 | | SpCas9 bound to FANCF off-target1 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target1 non-target strand, FANCF off-target1 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QR8
 
 | | SpCas9 bound to PTPRC off-target1 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQP
 
 | | SpCas9 bound to AAVS1 off-target3 DNA substrate | | Descriptor: | 1,2-ETHANEDIOL, AAVS1 off-target3 non-target strand, AAVS1 off-target3 target strand, ... | | Authors: | Pacesa, M, JInek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQS
 
 | | SpCas9 bound to FANCF on-target DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF on-target non-target strand, FANCF on-target target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQX
 
 | | SpCas9 bound to FANCF off-target5 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target5 non-target strand, FANCF off-target5 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QR0
 
 | | SpCas9 bound to TRAC off-target1 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQZ
 
 | | SpCas9 bound to FANCF off-target7 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target7 non-target strand, FANCF off-target7 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QQW
 
 | | SpCas9 bound to FANCF off-target4 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, FANCF off-target4 non-target strand, FANCF off-target4 target strand, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QR1
 
 | | SpCas9 bound to TRAC off-target2 DNA substrate | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
7QR7
 
 | | SpCas9 bound to AAVS1 off-target2 DNA substrate | | Descriptor: | AAVS1 off-target2 non-target strand, AAVS1 off-target2 target strand, AAVS1 sgRNA, ... | | Authors: | Pacesa, M, JInek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
