6S1W
 
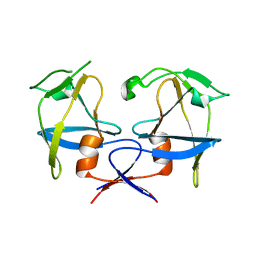 | | Crystal structure of dimeric M-PMV protease D26N mutant | | Descriptor: | Gag-Pro-Pol polyprotein | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S1V
 
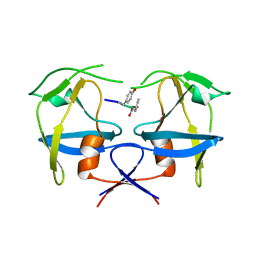 | | Crystal structure of dimeric M-PMV protease D26N mutant in complex with inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1EQT
 
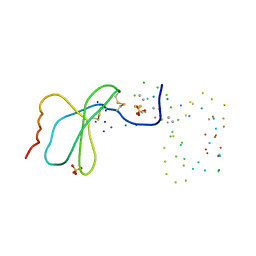 | | MET-RANTES | | Descriptor: | SULFATE ION, T-CELL SPECIFIC RANTES PROTEIN | | Authors: | Hoover, D.M, Shaw, J, Gryczynski, Z, Proudfoot, A.E.I, Wells, T. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of MET-RANTES: Comparison with Native RANTES and AOP-RANTES
PROTEIN PEPT.LETT., 7, 2000
|
|
3SQF
 
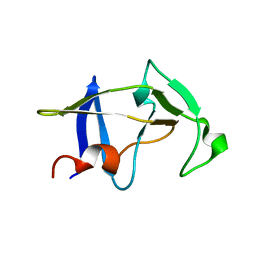 | | Crystal structure of monomeric M-PMV retroviral protease | | Descriptor: | Protease | | Authors: | Jaskolski, M, Kazmierczyk, M, Gilski, M, Krzywda, S, Pichova, I, Zabranska, H, Khatib, F, DiMaio, F, Cooper, S, Thompson, J, Popovic, Z, Baker, D, Group, Foldit Contenders | | Deposit date: | 2011-07-05 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6324 Å) | | Cite: | Crystal structure of a monomeric retroviral protease solved by protein folding game players.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4PHV
 
 | | X-RAY CRYSTAL STRUCTURE OF THE HIV PROTEASE COMPLEX WITH L-700,417, AN INHIBITOR WITH PSEUDO C2 SYMMETRY | | Descriptor: | HIV-1 PROTEASE, N,N-BIS(2-HYDROXY-1-INDANYL)-2,6- DIPHENYLMETHYL-4-HYDROXY-1,7-HEPTANDIAMIDE | | Authors: | Bone, R. | | Deposit date: | 1991-10-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Crystal Structure of the HIV Protease Complex with L-700,417, an Inhibitor with Pseudo C2 Symmetry
J.Am.Chem.Soc., 113, 1991
|
|
6KMP
 
 | | 100K X-ray structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate mimic KVS-1 | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, Protease | | Authors: | Das, A, Kovalevsky, A. | | Deposit date: | 2019-07-31 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
2J9K
 
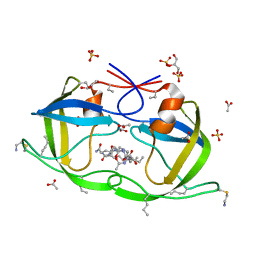 | | Atomic-resolution Crystal Structure of Chemically-Synthesized HIV-1 Protease Complexed with Inhibitor MVT-101 | | Descriptor: | ACETATE ION, GLYCEROL, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, ... | | Authors: | Malito, E, Shen, Y, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2006-11-11 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
J.Mol.Biol., 373, 2007
|
|
2J9J
 
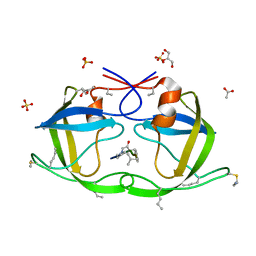 | | Atomic-resolution Crystal Structure of Chemically-Synthesized HIV-1 Protease Complexed with Inhibitor JG-365 | | Descriptor: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | Authors: | Malito, E, Shen, Y, Johnson, E.C.B, Tang, W.J. | | Deposit date: | 2006-11-11 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
J.Mol.Biol., 373, 2007
|
|
6PU8
 
 | | Room temperature X-ray structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate of keto-darunavir | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3,3-dihydroxy-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Kovalevsky, A, Das, A. | | Deposit date: | 2019-07-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
6PTP
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate mimic KVS-1 | | Descriptor: | HIV-1 Protease, N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide | | Authors: | Kovalevsky, A, Das, A. | | Deposit date: | 2019-07-16 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
