3CRO
 
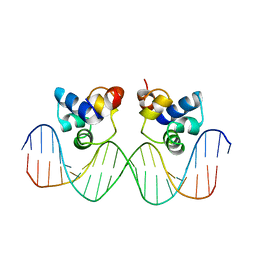 | | THE PHAGE 434 CRO/OR1 COMPLEX AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*AP*AP*GP*TP*AP*CP*AP*AP*AP*CP*TP*TP*TP*CP*TP*TP*G P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*GP*AP*AP*AP*GP*TP*TP*TP*GP*T P*AP*CP*T)-3'), PROTEIN (434 CRO) | | Authors: | Mondragon, A, Harrison, S.C. | | Deposit date: | 1990-07-06 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The phage 434 Cro/OR1 complex at 2.5 A resolution.
J.Mol.Biol., 219, 1991
|
|
6OUA
 
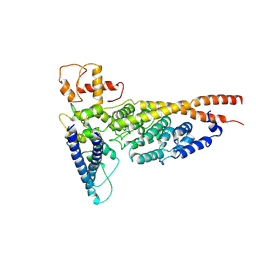 | | Cryo-EM structure of the yeast Ctf3 complex | | Descriptor: | Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, Inner kinetochore subunit MCM22 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2019-05-04 | | Release date: | 2019-05-15 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | The structure of the yeast Ctf3 complex.
Elife, 8, 2019
|
|
4HK3
 
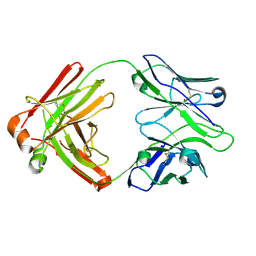 | | I2 Fab (unbound) from CH65-CH67 Lineage | | Descriptor: | I2 heavy chain, I2 light chain | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-14 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HKX
 
 | | Influenza hemagglutinin in complex with CH67 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH67 heavy chain, CH67 light chain, ... | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-15 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HK0
 
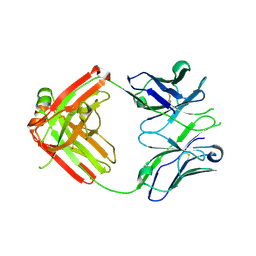 | | UCA Fab (unbound) from CH65-CH67 Lineage | | Descriptor: | UCA heavy chain, UCA light chain | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-14 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5F6I
 
 | |
5F6J
 
 | |
5F6H
 
 | |
4HKB
 
 | | CH67 Fab (unbound) from the CH65-67 Lineage | | Descriptor: | CH67 heavy chain, CH67 light chain | | Authors: | Schmidt, A.G, Harrison, S.C. | | Deposit date: | 2012-10-15 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4JE3
 
 | |
4IT3
 
 | | Crystal Structure of Iml3 from S. cerevisiae | | Descriptor: | Central kinetochore subunit IML3 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2013-01-17 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | An iml3-chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation.
Cell Rep, 5, 2013
|
|
1ZXQ
 
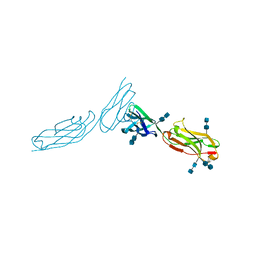 | | THE CRYSTAL STRUCTURE OF ICAM-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, INTERCELLULAR ADHESION MOLECULE-2 | | Authors: | Casasnovas, J.M, Springer, T.A, Harrison, S.C, Wang, J.-H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of ICAM-2 reveals a distinctive integrin recognition surface.
Nature, 387, 1997
|
|
3KZ4
 
 | | Crystal Structure of the Rotavirus Double Layered Particle | | Descriptor: | Inner capsid protein VP2, Intermediate capsid protein VP6, ZINC ION | | Authors: | Mcclain, B, Settembre, E.C, Bellamy, A.R, Harrison, S.C. | | Deposit date: | 2009-12-07 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | X-ray crystal structure of the rotavirus inner capsid particle at 3.8 A resolution.
J.Mol.Biol., 397, 2010
|
|
3ML6
 
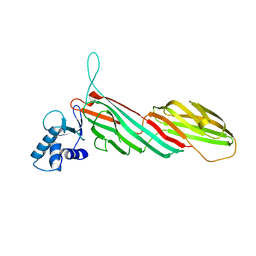 | | a complex between Dishevelled2 and clathrin adaptor AP-2 | | Descriptor: | Chimeric complex between protein Dishevelled2 homolog dvl-2 and clathrin adaptor AP-2 complex subunit mu | | Authors: | Yu, A, Xing, Y, Harrison, S.C, Kirchhausen, T.L. | | Deposit date: | 2010-04-16 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural analysis of the interaction between Dishevelled2 and clathrin AP-2 adaptor, a critical step in noncanonical Wnt signaling.
Structure, 18, 2010
|
|
3N7N
 
 | | Structure of Csm1/Lrs4 complex | | Descriptor: | Monopolin complex subunit CSM1, Monopolin complex subunit LRS4 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N4R
 
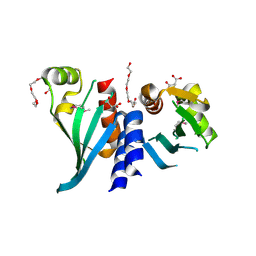 | | Structure of Csm1 C-terminal domain, R3 form | | Descriptor: | MALONATE ION, Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
2AJF
 
 | | Structure of SARS coronavirus spike receptor-binding domain complexed with its receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme-Related Carboxypeptidase (Ace2), CHLORIDE ION, ... | | Authors: | Li, F, Li, W, Farzan, M, Harrison, S.C. | | Deposit date: | 2005-08-01 | | Release date: | 2005-09-20 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of SARS coronavirus spike receptor-binding domain complexed with receptor.
Science, 309, 2005
|
|
3N4X
 
 | | Structure of Csm1 full-length | | Descriptor: | Monopolin complex subunit CSM1 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-23 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
2B6O
 
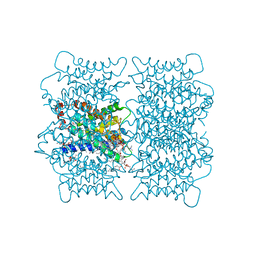 | | Electron crystallographic structure of lens Aquaporin-0 (AQP0) (lens MIP) at 1.9A resolution, in a closed pore state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.9 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
3N4S
 
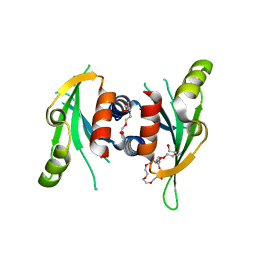 | | Structure of Csm1 C-terminal domain, P21212 form | | Descriptor: | Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
2B6P
 
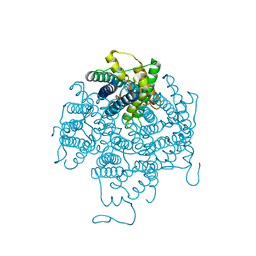 | | X-ray structure of lens Aquaporin-0 (AQP0) (lens MIP) in an open pore state | | Descriptor: | Lens fiber major intrinsic protein | | Authors: | Gonen, T, Cheng, Y, Sliz, P, Hiroaki, Y, Fujiyoshi, Y, Harrison, S.C, Walz, T. | | Deposit date: | 2005-10-03 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipid-protein interactions in double-layered two-dimensional AQP0 crystals.
Nature, 438, 2005
|
|
2CSE
 
 | | Features of Reovirus Outer-Capsid Protein mu1 Revealed by Electron and Image Reconstruction of the virion at 7.0-A Resolution | | Descriptor: | Minor core protein lambda 3, Sigma 2 protein, guanylyltransferase, ... | | Authors: | Zhang, X, Ji, Y, Zhang, L, Harrison, S.C, Marinescu, D.C, Nibert, M.L, Baker, T.S. | | Deposit date: | 2005-05-21 | | Release date: | 2005-10-18 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Features of reovirus outer capsid protein mu1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0 Angstrom resolution.
Structure, 13, 2005
|
|
1A02
 
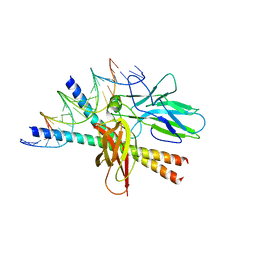 | | STRUCTURE OF THE DNA BINDING DOMAINS OF NFAT, FOS AND JUN BOUND TO DNA | | Descriptor: | AP-1 FRAGMENT FOS, AP-1 FRAGMENT JUN, DNA (5'-D(*DAP*DAP*DCP*DTP*DAP*DTP*DGP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DTP*DTP*DTP*DTP*DCP*DC)-3'), ... | | Authors: | Chen, L, Glover, J.N.M, Hogan, P.G, Rao, A, Harrison, S.C. | | Deposit date: | 1997-12-08 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA.
Nature, 392, 1998
|
|
1BGW
 
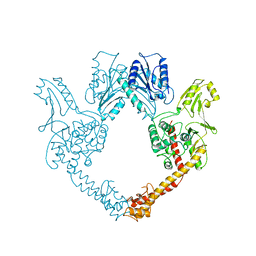 | | TOPOISOMERASE RESIDUES 410-1202, | | Descriptor: | TOPOISOMERASE | | Authors: | Berger, J.M, Gamblin, S.J, Harrison, S.C, Wang, J.C. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of DNA topoisomerase II.
Nature, 379, 1996
|
|
1BPO
 
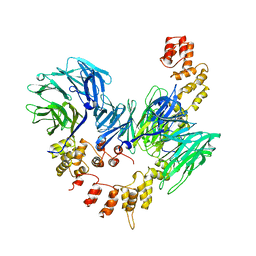 | | CLATHRIN HEAVY-CHAIN TERMINAL DOMAIN AND LINKER | | Descriptor: | PROTEIN (CLATHRIN) | | Authors: | Harr, E.T, Musacchio, A, Harrison, S.C, Kirchhausen, T. | | Deposit date: | 1998-08-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Atomic structure of clathrin: a beta propeller terminal domain joins an alpha zigzag linker.
Cell(Cambridge,Mass.), 95, 1998
|
|
