3UK3
 
 | | Crystal structure of ZNF217 bound to DNA | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', CHLORIDE ION, ... | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2011-11-08 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rediscovering DNA recognition by classical Zinc Fingers.
To be Published
|
|
3MJM
 
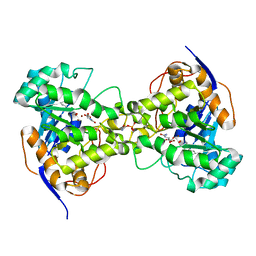 | | His257Ala mutant of dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Ernberg, K.E, Guss, J.M, Lee, M, Maher, M.J. | | Deposit date: | 2010-04-13 | | Release date: | 2011-03-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | His257Ala mutant of dihydroorotase from E. coli
To be Published
|
|
1FN2
 
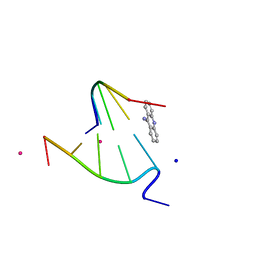 | | 9-AMINO-(N-(2-DIMETHYLAMINO)BUTYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CGTACG)2 | | Descriptor: | 9-AMINO-(N-(2-DIMETHYLAMINO)BUTYL)ACRIDINE-4-CARBOXAMIDE, COBALT (II) ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2000-08-19 | | Release date: | 2000-10-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel form of intercalation involving four DNA duplexes in an acridine-4-carboxamide complex of d(CGTACG)(2).
Nucleic Acids Res., 28, 2000
|
|
3OEP
 
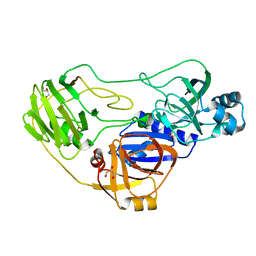 | | Crystal structure of TTHA0988 in space group P43212 | | Descriptor: | GLYCEROL, Putative uncharacterized protein TTHA0988, SULFATE ION | | Authors: | Jacques, D.A, Langley, D.B, Trewhella, J, Guss, J.M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-08-13 | | Release date: | 2010-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OPF
 
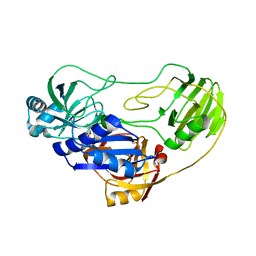 | | Crystal structure of TTHA0988 in space group P212121 | | Descriptor: | GLYCEROL, Putative uncharacterized protein TTHA0988, SULFATE ION | | Authors: | Jacques, D.A, Kuramitsu, S, Yokoyama, S, Trewhella, J, Guss, J.M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-01 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OVU
 
 | | Crystal Structure of Human Alpha-Haemoglobin Complexed with AHSP and the First NEAT Domain of IsdH from Staphylococcus aureus | | Descriptor: | Alpha-hemoglobin-stabilizing protein, Hemoglobin subunit alpha, Iron-regulated surface determinant protein H, ... | | Authors: | Jacques, D.A, Krishna Kumar, K, Caradoc-Davies, T.T, Langley, D.B, Mackay, J.P, Guss, J.M, Gell, D.A. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | A new haem pocket structure in alpha-haemoglobin
To be Published
|
|
1FN1
 
 | | CRYSTAL STRUCTURE OF 9-AMINO-(N-(2-DIMETHYLAMINO)BUTYL)ACRIDINE-4-CARBOXAMIDE BOUND TO D(CG(5BR)UACG)2 | | Descriptor: | 9-AMINO-(N-(2-DIMETHYLAMINO)BUTYL)ACRIDINE-4-CARBOXAMIDE, COBALT (II) ION, DNA (5'-D(*CP*GP*(BRO)UP*AP*CP*G)-3'), ... | | Authors: | Adams, A, Guss, J.M, Collyer, C.A, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2000-08-19 | | Release date: | 2000-10-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel form of intercalation involving four DNA duplexes in an acridine-4-carboxamide complex of d(CGTACG)(2).
Nucleic Acids Res., 28, 2000
|
|
3FYR
 
 | | Crystal structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis | | Descriptor: | Sporulation inhibitor sda | | Authors: | Jacques, D.A, Streamer, M, King, G.F, Guss, J.M, Trewhella, J, Langley, D.B. | | Deposit date: | 2009-01-23 | | Release date: | 2009-06-23 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the sporulation histidine kinase inhibitor Sda from Bacillus subtilis and insights into its solution state
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3ORE
 
 | | Crystal structure of TTHA0988 in space group P6522 | | Descriptor: | Putative uncharacterized protein TTHA0988 | | Authors: | Jacques, D.A, Kuramitsu, S, Yokoyama, S, Trewhella, J, Guss, J.M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-07 | | Release date: | 2011-02-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of TTHA0988 from Thermus thermophilus, a KipI-KipA homologue incorrectly annotated as an allophanate hydrolase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3PGB
 
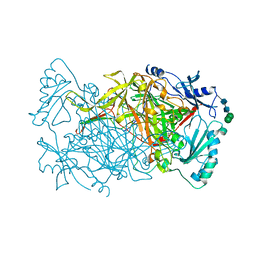 | | Crystal structure of Aspergillus nidulans amine oxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | McGrath, A.P, Guss, J.M. | | Deposit date: | 2010-11-01 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and Activity of Aspergillus nidulans Copper Amine Oxidase
Biochemistry, 50, 2011
|
|
3HI7
 
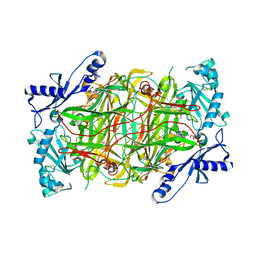 | | Crystal structure of human diamine oxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive amine oxidase, CALCIUM ION, ... | | Authors: | McGrath, A.P, Guss, J.M. | | Deposit date: | 2009-05-19 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structure and inhibition of human diamine oxidase
Biochemistry, 48, 2009
|
|
3HIG
 
 | |
3HII
 
 | |
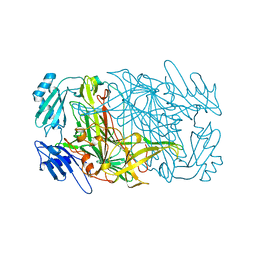
1AV4
 
 | |
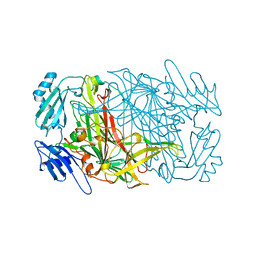
1AVL
 
 | |
1B2J
 
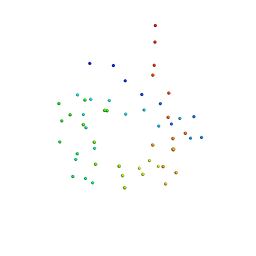 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-27 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1A16
 
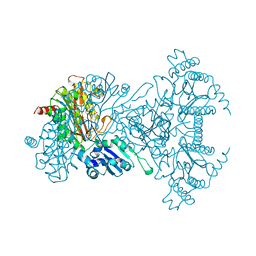 | | AMINOPEPTIDASE P FROM E. COLI WITH THE INHIBITOR PRO-LEU | | Descriptor: | AMINOPEPTIDASE P, LEUCINE, MANGANESE (II) ION, ... | | Authors: | Wilce, M.C, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BIK
 
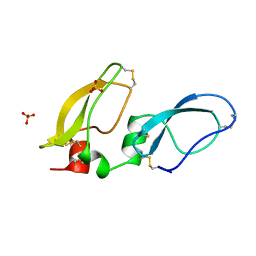 | | X-RAY STRUCTURE OF BIKUNIN FROM THE HUMAN INTER-ALPHA-INHIBITOR COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIKUNIN, SULFATE ION | | Authors: | Xu, Y, Carr, P.D, Guss, J.M, Ollis, D.L. | | Deposit date: | 1997-11-26 | | Release date: | 1999-03-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of bikunin from the inter-alpha-inhibitor complex: a serine protease inhibitor with two Kunitz domains.
J.Mol.Biol., 276, 1998
|
|
1BE7
 
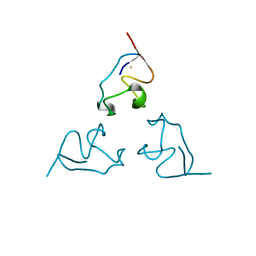 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN C42S MUTANT | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Maher, M, Guss, J.M, Wilce, M, Wedd, A.G. | | Deposit date: | 1998-05-20 | | Release date: | 1998-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Rubredoxin from Clostridium Pasteurianum: Mutation of the Iron Cysteinyl Ligands to Serine. Crystal and Molecular Structures of the Oxidised and Dithionite-Treated Forms of the Cys42Ser Mutant
J.Am.Chem.Soc., 120, 1998
|
|
1B13
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-26 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B2O
 
 | | CLOSTRIDIUM PASTEURIANUM RUBREDOXIN G10VG43A MUTANT | | Descriptor: | FE (III) ION, PROTEIN (RUBREDOXIN) | | Authors: | Maher, M.J, Guss, J.M, Wilce, M.C.J, Wedd, A.G. | | Deposit date: | 1998-11-30 | | Release date: | 1999-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rubredoxin from Clostridium pasteurianum. Structures of G10A, G43A and G10VG43A mutant proteins. Mutation of conserved glycine 10 to valine causes the 9-10 peptide link to invert.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1AVK
 
 | | CRYSTAL STRUCTURES OF THE COPPER-CONTAINING AMINE OXIDASE FROM ARTHROBACTER GLOBIFORMIS IN THE HOLO-AND APO-FORMS: IMPLICATIONS FOR THE BIOGENESIS OF TOPA QUINONE | | Descriptor: | AMINE OXIDASE | | Authors: | Wilce, M.C.J, Guss, J.M, Freeman, H.C, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 1997-09-16 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the copper-containing amine oxidase from Arthrobacter globiformis in the holo and apo forms: implications for the biogenesis of topaquinone.
Biochemistry, 36, 1997
|
|
1BAW
 
 | | PLASTOCYANIN FROM PHORMIDIUM LAMINOSUM | | Descriptor: | COPPER (II) ION, PLASTOCYANIN, ZINC ION | | Authors: | Bond, C.S, Guss, J.M, Freeman, H.C, Wagner, M.J, Howe, C.J, Bendall, D.S. | | Deposit date: | 1998-04-19 | | Release date: | 1999-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of plastocyanin from the cyanobacterium Phormidium laminosum.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2Z26
 
 | | Thr110Ala dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, MALONATE ION, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
2Z28
 
 | | Thr109Val dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
