6SJV
 
 | | Structure of HPV18 E6 oncoprotein in complex with mutant E6AP LxxLL motif | | Descriptor: | Maltodextrin-binding protein,Protein E6,Ubiquitin-protein ligase E3A, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Kostmann, C, Mitschler, A, Podjarny, A, Trave, G. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | Cellular target recognition by HPV18 and HPV49 oncoproteins
To be published
|
|
6SJA
 
 | | Structure of HPV16 E6 oncoprotein in complex with IRF3 LxxLL motif | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Interferon regulatory factor 3, Protein E6, ZINC ION, ... | | Authors: | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Mitschler, A, Podjarny, A, Trave, G. | | Deposit date: | 2019-08-13 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Deciphering de molecular and structural interaction between IRF3 and HPV16 E6
To be published
|
|
6SMV
 
 | | Structure of HPV49 E6 protein in complex with MAML1 LxxLL motif | | Descriptor: | DI(HYDROXYETHYL)ETHER, Maltose/maltodextrin-binding periplasmic protein,Protein E6,Mastermind-like protein 1, ZINC ION, ... | | Authors: | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Kostmann, C, Mitschler, A, Podjarny, A, Trave, G. | | Deposit date: | 2019-08-22 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Cellular target recognition by HPV18 and HPV49 oncoproteins
To be published
|
|
6SIV
 
 | | Structure of HPV16 E6 oncoprotein in complex with mutant IRF3 LxxLL motif | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Interferon regulatory factor 3, Protein E6, ZINC ION, ... | | Authors: | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Mitschler, A, Podjarny, A, Trave, G. | | Deposit date: | 2019-08-12 | | Release date: | 2019-08-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Deciphering the molecular and structural interaction between IRF3 and HPV16 E6
To be published
|
|
5NRA
 
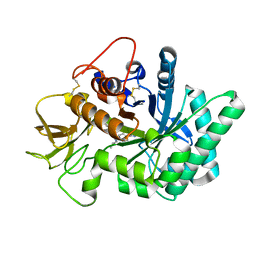 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 7g | | Descriptor: | 1-(5-azanyl-4~{H}-1,2,4-triazol-3-yl)-~{N}-[2-(4-bromophenyl)ethyl]-~{N}-(2-methylpropyl)piperidin-4-amine, Chitotriosidase-1, GLYCEROL | | Authors: | Mazur, M, Olczak, J, Olejniczak, S, Koralewski, R, Czestkowski, W, Jedrzejczak, A, Golab, J, Dzwonek, K, Dymek, B, Sklepkiewicz, P, Zagozdzon, A, Noonan, T, Mahboubi, K, Conway, B, Sheeler, R, Beckett, P, Hungerford, W.M, Podjarny, A, Mitschler, A, Cousido-Siah, A, Fadel, F, Golebiowski, A. | | Deposit date: | 2017-04-22 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.267 Å) | | Cite: | Targeting Acidic Mammalian chitinase Is Effective in Animal Model of Asthma.
J. Med. Chem., 61, 2018
|
|
6TWX
 
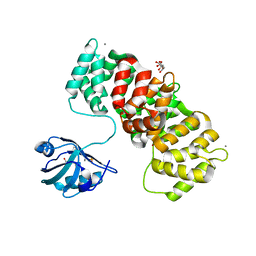 | | MAGI1_2 complexed with a phosphorylated 16E6 peptide | | Descriptor: | 16E6 peptide, CALCIUM ION, CITRIC ACID, ... | | Authors: | Gogl, G, Cousido-Siah, A, Trave, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dual Specificity PDZ- and 14-3-3-Binding Motifs: A Structural and Interactomics Study.
Structure, 28, 2020
|
|
6SLM
 
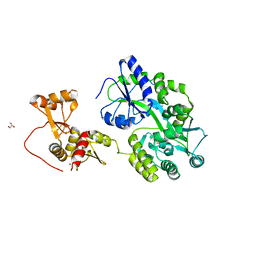 | | Crystal structure of full-length HPV31 E6 oncoprotein in complex with LXXLL peptide of ubiquitin ligase E6AP | | Descriptor: | GLYCEROL, Maltose/maltodextrin-binding periplasmic protein,Protein E6,Ubiquitin-protein ligase E3A, ZINC ION, ... | | Authors: | Conrady, M, Gogl, G, Cousido-Siah, A, Mitschler, A, Trave, G, Simon, C. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of High-Risk Papillomavirus 31 E6 Oncogenic Protein and Characterization of E6/E6AP/p53 Complex Formation.
J.Virol., 95, 2020
|
|
5M2F
 
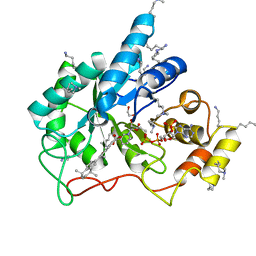 | | Crystal structure of human AKR1B10 complexed with NADP+ and the synthetic retinoid UVI2008 | | Descriptor: | 1,2-ETHANEDIOL, 3-bromo-4-[(1E)-2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)prop-1-en-1-yl]benzoic acid, Aldo-keto reductase family 1 member B10, ... | | Authors: | Ruiz, F.X, Cousido-Siah, A, Mitschler, A, Porte, S, Alvarez, S, Dominguez, M, Alvarez, R, de Lera, A.R, Pares, X, Farres, J, Podjarny, A. | | Deposit date: | 2016-10-12 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural basis for the inhibition of AKR1B10 by the C3 brominated TTNPB derivative UVI2008.
Chem. Biol. Interact., 276, 2017
|
|
6SQC
 
 | | Crystal structure of complex between nuclear coactivator binding domain of CBP and [1040-1086]ACTR containing alpha-methylated Leu1055 and Leu1076 | | Descriptor: | 1,2-ETHANEDIOL, Maltose/maltodextrin-binding periplasmic protein,CREB-binding protein, Nuclear receptor coactivator 3, ... | | Authors: | Bauer, V, Schmidtgall, B, Gogl, G, Dolenc, j, Osz, J, Kostmann, C, Mitschler, A, Cousido-Siah, A, Rochel, N, Trave, G, Kieffer, B, Torbeev, V. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Conformational editing of intrinsically disordered protein by alpha-methylation.
Chem Sci, 12, 2020
|
|
4NY6
 
 | | Neutron structure of leucine and valine methyl protonated type III antifreeze | | Descriptor: | Type-3 ice-structuring protein HPLC 12 | | Authors: | Fisher, S.J, Blakeley, M.P, Howard, E.I, Petite-Haertlein, I, Haertlein, M, Mitschler, A, Cousido-Siah, A, Salvaya, A.G, Popov, A, Muller-Dieckmann, C, Petrova, T, Podjarny, A.D. | | Deposit date: | 2013-12-10 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (1.05 Å), X-RAY DIFFRACTION | | Cite: | Perdeuteration: improved visualization of solvent structure in neutron macromolecular crystallography.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LP1
 
 | | Crystal structure of CyaY protein from Psychromonas ingrahamii in complex with Eu(III) | | Descriptor: | EUROPIUM (III) ION, Protein CyaY | | Authors: | Noguera, M.E, Roman, E.A, Cousido-Siah, A, Mitschler, A, Podjarny, A, Santos, J. | | Deposit date: | 2013-07-14 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural characterization of metal binding to a cold-adapted frataxin
J.Biol.Inorg.Chem., 20, 2015
|
|
4LK8
 
 | | Crystal structure of CyaY protein from Psychromonas ingrahamii in complex with Co(II) | | Descriptor: | COBALT (II) ION, Protein CyaY | | Authors: | Noguera, M.E, Roman, E.A, Cousido-Siah, A, Mitschler, A, Podjarny, A, Santos, J. | | Deposit date: | 2013-07-07 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | Structural characterization of metal binding to a cold-adapted frataxin
J.Biol.Inorg.Chem., 20, 2015
|
|
3MU5
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. Third step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MTY
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. First step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MU1
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. Fifth step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3ODF
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. Second step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-08-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3ODD
 
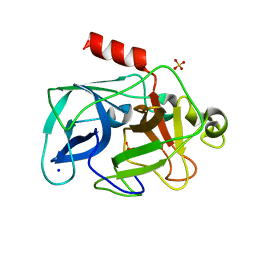 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. Second step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-08-11 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4WKH
 
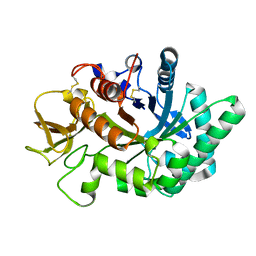 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (1mM) at 1.05 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WKF
 
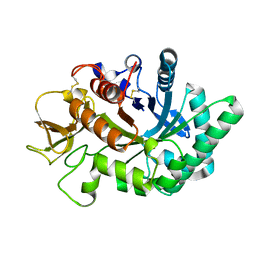 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (2.5mM) at 1.10 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WKA
 
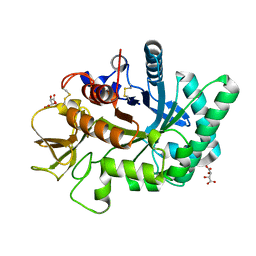 | | Crystal structure of human chitotriosidase-1 catalytic domain at 0.95 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3MU0
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region A of the crystal. Third step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MU8
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. Fifth step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-02 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MU4
 
 | | Comparison of the character and the speed of X-ray-induced structural changes of porcine pancreatic elastase at two temperatures, 100 and 15K. The data set was collected from region B of the crystal. First step of radiation damage | | Descriptor: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | Authors: | Petrova, T, Ginell, S, Mitschler, A, Cousido-Siah, A, Hazemann, I, Podjarny, A, Joachimiak, A. | | Deposit date: | 2010-05-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7Q46
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of pericentriolar material 1 protein | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Pericentriolar material 1 protein | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46002531 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of pericentriolar material 1 protein
To Be Published
|
|
7Q43
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of dedicator of cytokinesis protein 10 | | Descriptor: | CITRIC ACID, Dedicator of cytokinesis protein 10 peptide, E3 ubiquitin-protein ligase HERC2 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.40002346 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of dedicator of cytokinesis protein 10
To Be Published
|
|
