6LNH
 
 | | Crystal structure of IDO from Bacillus thuringiensis | | Descriptor: | FE (III) ION, L-isoleucine-4-hydroxylase, MERCURY (II) ION | | Authors: | Feng, Y, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of IDO from Bacillus thuringiensis
to be published
|
|
7XWJ
 
 | | structure of patulin-detoxifying enzyme Y155F with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWI
 
 | | structure of patulin-detoxifying enzyme with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWH
 
 | | structure of patulin-detoxifying enzyme with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWL
 
 | | structure of patulin-detoxifying enzyme Y155F/V187F with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWN
 
 | | structure of patulin-detoxifying enzyme Y155F/V187K with NADPH and substrate | | Descriptor: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
7XWK
 
 | | structure of patulin-detoxifying enzyme Y155F with NADPH and substrate | | Descriptor: | (4~{S})-4-oxidanyl-4,6-dihydrofuro[3,2-c]pyran-2-one, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase | | Authors: | Dai, L, Li, H, Hu, Y, Guo, R.T, Chen, C.C. | | Deposit date: | 2022-05-26 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-based rational design of a short-chain dehydrogenase/reductase for improving activity toward mycotoxin patulin.
Int.J.Biol.Macromol., 222, 2022
|
|
4TSR
 
 | |
5I78
 
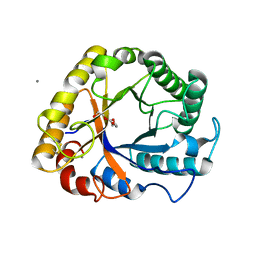 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | Authors: | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
5I79
 
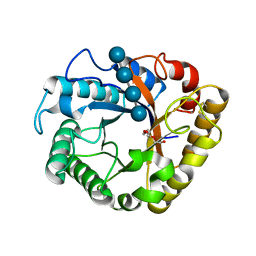 | | Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | Authors: | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
5I77
 
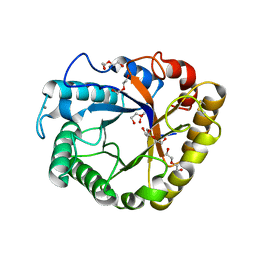 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, Y.J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
1DIK
 
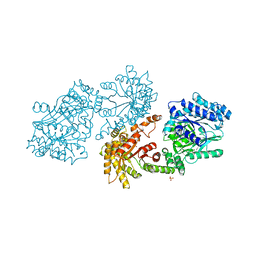 | | PYRUVATE PHOSPHATE DIKINASE | | Descriptor: | PYRUVATE PHOSPHATE DIKINASE, SULFATE ION | | Authors: | Herzberg, O, Chen, C.C.H. | | Deposit date: | 1995-12-06 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
3VNI
 
 | | Crystal structures of D-Psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars | | Descriptor: | MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3W01
 
 | | Crystal structure of PcrB complexed with PEG from Staphylococcus aureus subsp. aureus Mu3 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, TRIETHYLENE GLYCOL | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VNJ
 
 | | Crystal structures of D-Psicose 3-epimerase with D-psicose from Clostridium cellulolyticum H10 | | Descriptor: | D-psicose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VZZ
 
 | | Crystal structure of PcrB complexed with FsPP from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VNM
 
 | | Crystal structures of D-Psicose 3-epimerase with D-sorbose from Clostridium cellulolyticum H10 | | Descriptor: | D-sorbose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-17 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VSV
 
 | | The complex structure of XylC with xylose | | Descriptor: | Xylosidase, alpha-D-xylopyranose, beta-D-xylopyranose | | Authors: | Huang, C.H, Sun, Y, Ko, T.P, Ma, Y, Chen, C.C, Zheng, Y, Chan, H.C, Pang, X, Wiegel, J, Shao, W, Guo, R.T. | | Deposit date: | 2012-05-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The substrate/product-binding modes of a novel GH120 beta-xylosidase (XylC) from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Biochem.J., 448, 2012
|
|
3VZX
 
 | | Crystal structure of PcrB from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VNK
 
 | | Crystal structures of D-Psicose 3-epimerase with D-fructose from Clostridium cellulolyticum H10 | | Descriptor: | D-fructose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VZY
 
 | | Crystal structure of PcrB complexed with G1P from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3VNL
 
 | | Crystal structures of D-Psicose 3-epimerase with D-tagatose from Clostridium cellulolyticum H10 | | Descriptor: | D-tagatose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3W00
 
 | | Crystal structure of PcrB complexed with G1P and FsPP from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, PHOSPHATE ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3WCI
 
 | | The complex structure of HsSQS wtih ligand,BPH1325 | | Descriptor: | Squalene synthase, hydrogen [(1R)-1-hydroxy-2-(3-pentadecyl-1H-imidazol-3-ium-1-yl)-1-phosphonoethyl]phosphonate | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
3WCB
 
 | | The complex structure of TcSQS with ligand, BPH1237 | | Descriptor: | Farnesyltransferase, putative, hydrogen [(1R)-2-(3-decyl-1H-imidazol-3-ium-1-yl)-1-hydroxy-1-phosphonoethyl]phosphonate | | Authors: | Shang, N, Li, Q, Ko, T.P, Chan, H.C, Huang, C.H, Ren, F, Zheng, Y, Zhu, Z, Chen, C.C, Guo, R.T. | | Deposit date: | 2013-05-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Squalene synthase as a target for Chagas disease therapeutics.
Plos Pathog., 10, 2014
|
|
