7DFS
 
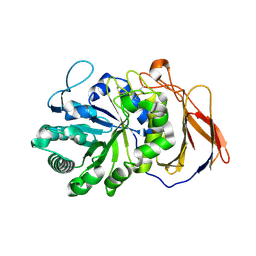 | | Crystal structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S - Rha-GlcA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase, alpha-D-mannopyranose, ... | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
7DFQ
 
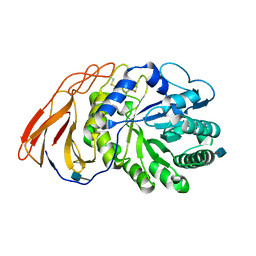 | | Crystal Structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S, ligand-free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
5BXP
 
 | | LNBase in complex with LNB-LOGNAc | | Descriptor: | Lacto-N-biosidase, SULFATE ION, beta-D-galactopyranose-(1-3)-N-acetylglucosaminono-1,5-lactone (Z)-oxime | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5BXS
 
 | | LNBase in complex with LNB-NHAcCAS | | Descriptor: | 6-ACETAMIDO-6-DEOXY-CASTANOSPERMINE, Lacto-N-biosidase, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5BXT
 
 | | LNBase in complex with LNB-NHAcAUS | | Descriptor: | Lacto-N-biosidase, N-{[(1R,2R,3R,7S,7aR)-1,2,7-trihydroxyhexahydro-1H-pyrrolizin-3-yl]methyl}acetamide, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
7BVT
 
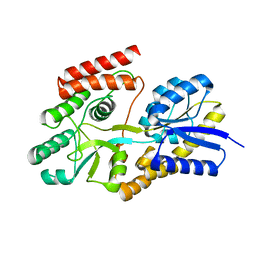 | | Crystal structure of cyclic alpha-maltosyl-1,6-maltose binding protein from Arthrobacter globiformis | | Descriptor: | Hypothetical sugar ABC-transporter sugar binding protein, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2020-04-11 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular analysis of cyclic alpha-maltosyl-(1→6)-maltose binding protein in the bacterial metabolic pathway.
Plos One, 15, 2020
|
|
5BXR
 
 | | LNBase in complex with LNB-NHAcDNJ | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, Lacto-N-biosidase, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5B0R
 
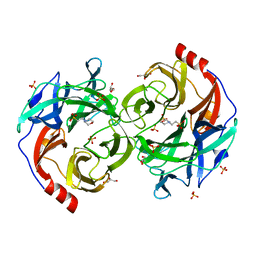 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - beta-1,2-mannobiose complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5B0P
 
 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - glycerol complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5B0S
 
 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - beta-1,2-mannotriose complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5ZNM
 
 | | Colicin D Central Domain and C-terminal tRNase domain | | Descriptor: | Colicin-D, GLYCEROL, SULFATE ION | | Authors: | Chang, J.W, Sato, Y, Ogawa, T, Arakawa, T, Fukai, S, Fushinobu, S, Masaki, H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the central and the C-terminal RNase domains of colicin D implicated its translocation pathway through inner membrane of target cell
J. Biochem., 164, 2018
|
|
5B0Q
 
 | | beta-1,2-Mannobiose phosphorylase from Listeria innocua - mannose complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lin0857 protein, SULFATE ION, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
7ESM
 
 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex | | Descriptor: | ACETATE ION, L-rhamnose-alpha-1,4-D-glucuronate lyase, SODIUM ION, ... | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
7ESN
 
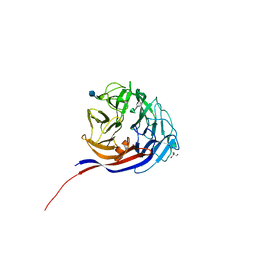 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, H105F Rha-GlcA complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, L-Rhamnose-alpha-1,4-D-glucuronate lyase, ... | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
7ESK
 
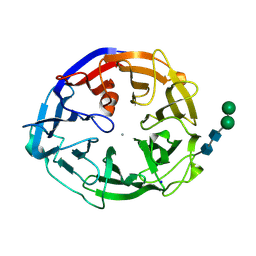 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, Ligand free form | | Descriptor: | CALCIUM ION, L-Rhamnose-alpha-1,4-D-glucuronate lyase, SODIUM ION, ... | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
7ESL
 
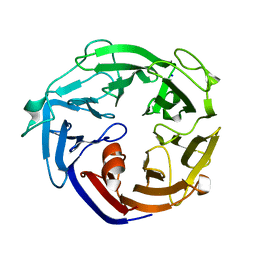 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, N247A N-glycan free form | | Descriptor: | L-rhamnose-alpha-1,4-D-glucuronate lyase, SODIUM ION | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
6A94
 
 | | Crystal structure of 5-HT2AR in complex with zotepine | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(3-chloranylbenzo[b][1]benzothiepin-5-yl)oxy-N,N-dimethyl-ethanamine, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Kimura, T.K, Asada, H, Inoue, A, Kadji, F.M.N, Im, D, Mori, C, Arakawa, T, Hirata, K, Nomura, Y, Nomura, N, Aoki, J, Iwata, S, Shimamura, T. | | Deposit date: | 2018-07-11 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the 5-HT2Areceptor in complex with the antipsychotics risperidone and zotepine.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6A93
 
 | | Crystal structure of 5-HT2AR in complex with risperidone | | Descriptor: | 3-[2-[4-(6-fluoranyl-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl]-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Kimura, T.K, Asada, H, Inoue, A, Kadji, F.M.N, Im, D, Mori, C, Arakawa, T, Hirata, K, Nomura, Y, Nomura, N, Aoki, J, Iwata, S, Shimamura, T. | | Deposit date: | 2018-07-11 | | Release date: | 2019-02-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the 5-HT2Areceptor in complex with the antipsychotics risperidone and zotepine.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4ZLF
 
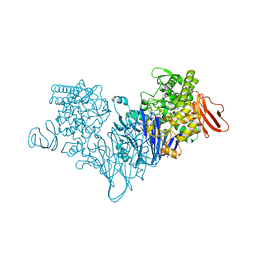 | | Cellobionic acid phosphorylase - cellobionic acid complex | | Descriptor: | 4-O-beta-D-glucopyranosyl-D-gluconic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZLE
 
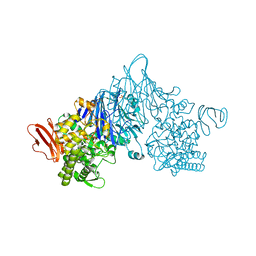 | | Cellobionic acid phosphorylase - ligand free structure | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZLG
 
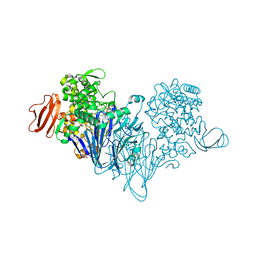 | | Cellobionic acid phosphorylase - gluconic acid complex | | Descriptor: | CHLORIDE ION, D-gluconic acid, D-glucono-1,5-lactone, ... | | Authors: | Nam, Y.W, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-05-01 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
5YSD
 
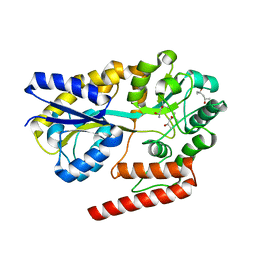 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotriose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSF
 
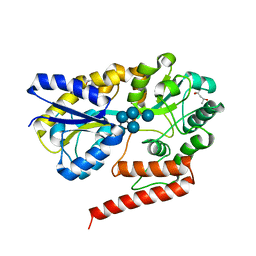 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophoropentaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSE
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotetraose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSB
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in ligand-free form | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lin1841 protein, ZINC ION | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
