4HYG
 
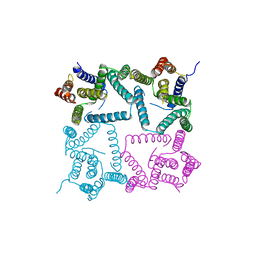 | | Structure of a presenilin family intramembrane aspartate protease in C222 space group | | Descriptor: | Putative uncharacterized protein | | Authors: | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
4HYC
 
 | | Structure of a presenilin family intramembrane aspartate protease in P2 space group | | Descriptor: | Putative uncharacterized protein | | Authors: | Li, X, Dang, S, Yan, C, Wang, J, Shi, Y. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of a presenilin family intramembrane aspartate protease
Nature, 493, 2013
|
|
4ITJ
 
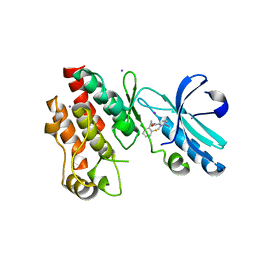 | | Crystal structure of RIP1 kinase in complex with necrostatin-4 | | Descriptor: | IODIDE ION, N-[(1S)-1-(2-chloro-6-fluorophenyl)ethyl]-5-cyano-1-methyl-1H-pyrrole-2-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Xie, T, Peng, W, Liu, Y, Yan, C, Shi, Y. | | Deposit date: | 2013-01-18 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of RIP1 Inhibition by Necrostatins.
Structure, 21, 2013
|
|
4ITI
 
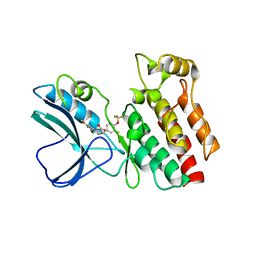 | | Crystal structure of RIP1 kinase in complex with necrostatin-3 analog | | Descriptor: | 1-{(3S,3aS)-3-[3-fluoro-4-(trifluoromethoxy)phenyl]-8-methoxy-3,3a,4,5-tetrahydro-2H-benzo[g]indazol-2-yl}-2-hydroxyethanone, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Xie, T, Peng, W, Liu, Y, Yan, C, Shi, Y. | | Deposit date: | 2013-01-18 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural Basis of RIP1 Inhibition by Necrostatins.
Structure, 21, 2013
|
|
4ITH
 
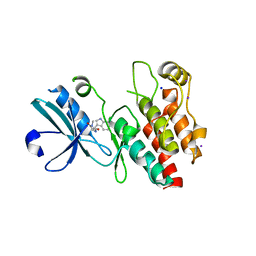 | | Crystal structure of RIP1 kinase in complex with necrostatin-1 analog | | Descriptor: | (5R)-5-[(7-chloro-1H-indol-3-yl)methyl]-3-methylimidazolidine-2,4-dione, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1, ... | | Authors: | Xie, T, Peng, W, Liu, Y, Yan, C, Shi, Y. | | Deposit date: | 2013-01-18 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of RIP1 Inhibition by Necrostatins.
Structure, 21, 2013
|
|
4M68
 
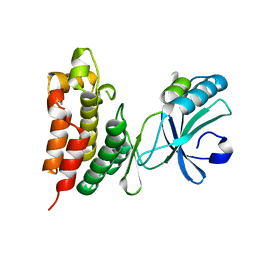 | | Crystal structure of the mouse MLKL kinase-like domain | | Descriptor: | GLYCEROL, Mixed lineage kinase domain-like protein | | Authors: | Xie, T, Peng, W, Yan, C, Wu, J, Shi, Y. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Structural Insights into RIP3-Mediated Necroptotic Signaling
Cell Rep, 5, 2013
|
|
5XJC
 
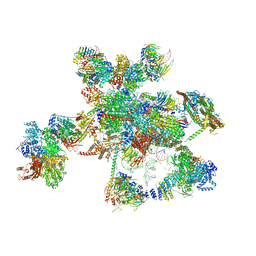 | | Cryo-EM structure of the human spliceosome just prior to exon ligation at 3.6 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, X, Yan, C, Hang, J, Finci, I.L, Lei, J, Shi, Y. | | Deposit date: | 2017-04-30 | | Release date: | 2017-07-05 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An Atomic Structure of the Human Spliceosome
Cell, 169, 2017
|
|
7CP9
 
 | | Cryo-EM structure of human mitochondrial translocase TOM complex at 3.0 angstrom. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, ... | | Authors: | Guan, Z, Yan, L, Wang, Q, Yan, C, Yin, P. | | Deposit date: | 2020-08-06 | | Release date: | 2021-04-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into assembly of human mitochondrial translocase TOM complex.
Cell Discov, 7, 2021
|
|
5YLZ
 
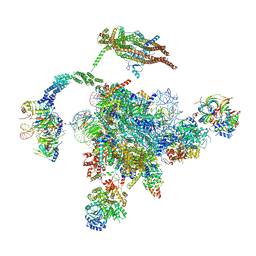 | | Cryo-EM Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae at 3.6 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae
Cell, 171, 2017
|
|
5Y88
 
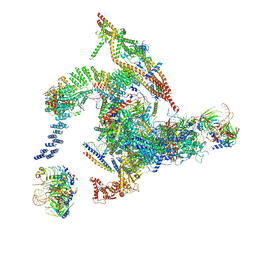 | | Cryo-EM structure of the intron-lariat spliceosome ready for disassembly from S.cerevisiae at 3.5 angstrom | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat, ... | | Authors: | Wan, R, Yan, C, Bai, R, Lei, J, Shi, Y. | | Deposit date: | 2017-08-20 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of an Intron Lariat Spliceosome from Saccharomyces cerevisiae
Cell(Cambridge,Mass.), 171, 2017
|
|
5YZG
 
 | | The Cryo-EM Structure of Human Catalytic Step I Spliceosome (C complex) at 4.1 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Lei, J, Shi, Y. | | Deposit date: | 2017-12-14 | | Release date: | 2018-08-08 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of a human catalytic step I spliceosome
Science, 359, 2018
|
|
5Z56
 
 | | cryo-EM structure of a human activated spliceosome (mature Bact) at 5.1 angstrom. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | Deposit date: | 2018-01-17 | | Release date: | 2018-09-19 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5Z58
 
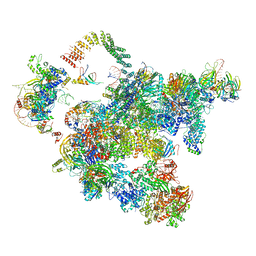 | | Cryo-EM structure of a human activated spliceosome (early Bact) at 4.9 angstrom. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | Deposit date: | 2018-01-17 | | Release date: | 2018-09-19 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
5Z57
 
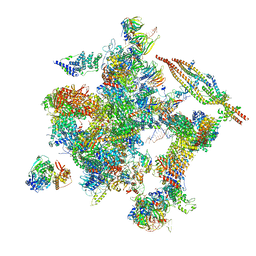 | | Cryo-EM structure of the human activated spliceosome (late Bact) at 6.5 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ALANINE, BUD13 homolog, ... | | Authors: | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | Deposit date: | 2018-01-17 | | Release date: | 2018-09-19 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
6M49
 
 | | cryo-EM structure of Scap/Insig complex in the present of 25-hydroxyl cholesterol. | | Descriptor: | 25-HYDROXYCHOLESTEROL, Insulin-induced gene 2 protein, Sterol regulatory element-binding protein cleavage-activating protein,Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Yan, R, Cao, P, Song, W, Qian, H, Du, X, Coates, H.W, Zhao, X, Li, Y, Gao, S, Gong, X, Liu, X, Sui, J, Lei, J, Yang, H, Brown, A.J, Zhou, Q, Yan, C, Yan, N. | | Deposit date: | 2020-03-06 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols.
Science, 371, 2021
|
|
5ZWO
 
 | | Cryo-EM structure of the yeast B complex at average resolution of 3.9 angstrom | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 23 kDa U4/U6.U5 small nuclear ribonucleoprotein component, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, ... | | Authors: | Bai, R, Wan, R, Yan, C, Shi, Y. | | Deposit date: | 2018-05-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
5ZWN
 
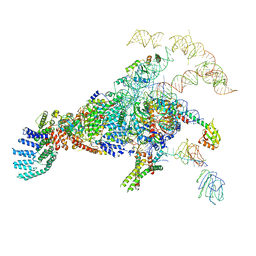 | | Cryo-EM structure of the yeast pre-B complex at an average resolution of 3.3 angstrom (Part II: U1 snRNP region) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, Pre-mRNA-processing factor 39, Pre-mRNA-splicing ATP-dependent RNA helicase PRP28, ... | | Authors: | Bai, R, Wan, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2018-05-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
5ZWM
 
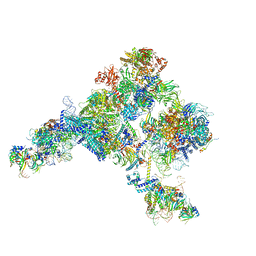 | | Cryo-EM structure of the yeast pre-B complex at an average resolution of 3.4~4.6 angstrom (tri-snRNP and U2 snRNP Part) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Cold sensitive U2 snRNA suppressor 1, ... | | Authors: | Bai, R, Wan, R, Yan, C, Lei, J, Shi, Y. | | Deposit date: | 2018-05-16 | | Release date: | 2018-08-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the fully assembledSaccharomyces cerevisiaespliceosome before activation
Science, 360, 2018
|
|
6AH0
 
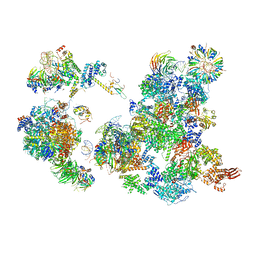 | | The Cryo-EM Structure of the Precusor of Human Pre-catalytic Spliceosome (pre-B complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
8JTZ
 
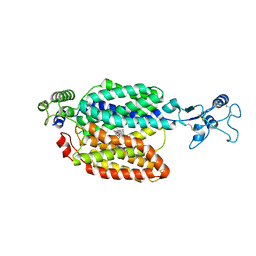 | | hOCT1 in complex with spironolactone in outward facing partially occluded conformation | | Descriptor: | SPIRONOLACTONE, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTT
 
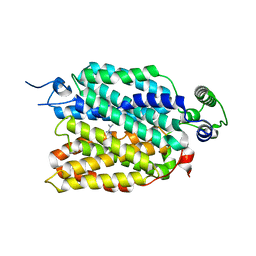 | | hOCT1 in complex with metformin in outward occluded conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTW
 
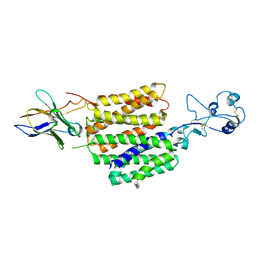 | | hOCT1 in complex with nb5660 in inward facing partially open 1 conformation | | Descriptor: | Solute carrier family 22 member 1, nanobody 56 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTS
 
 | | hOCT1 in complex with metformin in outward open conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTX
 
 | | hOCT1 in complex with nb5660 in inward facing fully open conformation | | Descriptor: | Solute carrier family 22 member 1, nanobody 56 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTY
 
 | | hOCT1 in complex with nb5660 in inward facing partially open 2 conformation | | Descriptor: | Solute carrier family 22 member 1, nanobody 56 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
