2H68
 
 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-30 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2H6K
 
 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-31 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3SOX
 
 | | Structure of UHRF1 PHD finger in the free form | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Rajakumara, E, Patel, D.J. | | Deposit date: | 2011-06-30 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6501 Å) | | Cite: | PHD Finger Recognition of Unmodified Histone H3R2 Links UHRF1 to Regulation of Euchromatic Gene Expression.
Mol.Cell, 43, 2011
|
|
2F6N
 
 | |
3U5O
 
 | |
2F8S
 
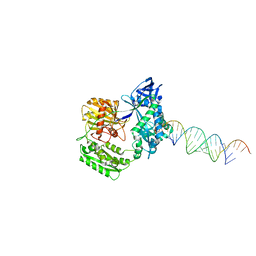 | | Crystal structure of Aa-Ago with externally-bound siRNA | | Descriptor: | 5'-R(P*AP*GP*AP*CP*AP*GP*CP*AP*UP*AP*UP*AP*UP*GP*CP*UP*GP*UP*CP*UP*UP*U)-3', Argonaute protein | | Authors: | Yuan, Y.R, Chen, H.Y, Patel, D.J. | | Deposit date: | 2005-12-03 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Potential Protein-RNA Recognition Event along the RISC-Loading Pathway from the Structure of A. aeolicus Argonaute with Externally Bound siRNA.
Structure, 14, 2006
|
|
3U5P
 
 | |
3U5N
 
 | |
2EUK
 
 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with 24:1 Galactosylceramide | | Descriptor: | (15E)-TETRACOS-15-ENOIC ACID, Glycolipid transfer protein, N-OCTANE, ... | | Authors: | Malinina, L, Malakhova, M.L, Kanack, A.T, Abagyan, R, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2EVT
 
 | | Crystal structure of D48V mutant of human Glycolipid Transfer Protein | | Descriptor: | Glycolipid transfer protein, HEXANE | | Authors: | Malinina, L, Malakhova, M.L, Teplov, A, Brown, R.E, Patel, D.J. | | Deposit date: | 2005-10-31 | | Release date: | 2005-11-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure.
Plos Biol., 4, 2006
|
|
2H6N
 
 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me2 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-31 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
3UIJ
 
 | |
3UIK
 
 | |
3UIH
 
 | |
5U33
 
 | | Crystal structure of AacC2c1-sgRNA-extended non-target DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease C2c1, Non-target DNA strand, SULFATE ION, ... | | Authors: | Yang, H, Gao, P, Rajashankar, K.R, Patel, D.J. | | Deposit date: | 2016-12-01 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | PAM-Dependent Target DNA Recognition and Cleavage by C2c1 CRISPR-Cas Endonuclease.
Cell, 167, 2016
|
|
5VW1
 
 | | Crystal structure of SpyCas9-sgRNA-AcrIIA4 ternary complex | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-05-21 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Inhibition Mechanism of an Anti-CRISPR Suppressor AcrIIA4 Targeting SpyCas9.
Mol. Cell, 67, 2017
|
|
4YB0
 
 | | 3',3'-cGAMP riboswitch bound with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
4YAZ
 
 | | 3',3'-cGAMP riboswitch bound with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
4YB1
 
 | | 20A Mutant c-di-GMP Vc2 Riboswitch bound with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, MAGNESIUM ION, RNA (91-MER), ... | | Authors: | Ren, A.M, Patel, D.J, Rajashankar, R.K. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Structural Basis for Molecular Discrimination by a 3',3'-cGAMP Sensing Riboswitch.
Cell Rep, 11, 2015
|
|
3CZ3
 
 | | Crystal structure of Tomato Aspermy Virus 2b in complex with siRNA | | Descriptor: | Protein 2b, RNA (5'-R(P*CP*GP*UP*AP*CP*GP*CP*GP*GP*AP*AP*UP*AP*CP*UP*UP*CP*GP*A)-3'), RNA (5'-R(P*UP*CP*GP*AP*AP*GP*UP*AP*UP*UP*CP*CP*GP*CP*GP*UP*AP*CP*G)-3') | | Authors: | Ma, J.B, Li, F, Ding, S.W, Patel, D.J. | | Deposit date: | 2008-04-27 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural Basis for siRNA Recognition by 2b, a Viral Suppressor of Non-Cell Autonomous RNA Silencing
To be Published
|
|
3D2N
 
 | |
3D2S
 
 | |
3D2Q
 
 | |
149D
 
 | |
136D
 
 | |
