3FF9
 
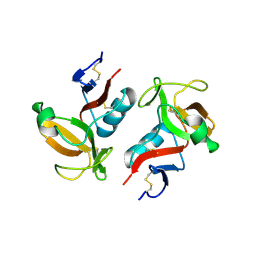 | | Structure of NK cell receptor KLRG1 | | Descriptor: | Killer cell lectin-like receptor subfamily G member 1 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
1VF6
 
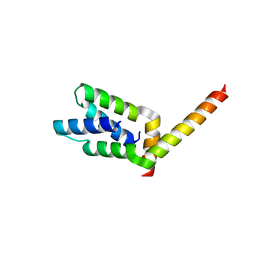 | | 2.1 Angstrom crystal structure of the PALS-1-L27N and PATJ L27 heterodimer complex | | Descriptor: | MAGUK p55 subfamily member 5, PALS1-associated tight junction protein | | Authors: | Li, Y, Lavie, A, Margolis, B, Karnak, D. | | Deposit date: | 2004-04-09 | | Release date: | 2004-04-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for L27 domain-mediated assembly of signaling and cell polarity complexes.
Embo J., 23, 2004
|
|
8EFN
 
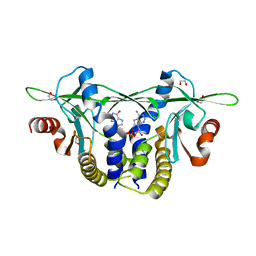 | | Structure of Sp-STING3 from Stylophora pistillata coral in complex with 3',3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon genes protein | | Authors: | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8EFM
 
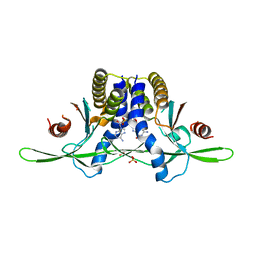 | | Structure of coral STING receptor from Stylophora pistillata in complex with 2',3'-cGAMP | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
2PJV
 
 | |
3FF8
 
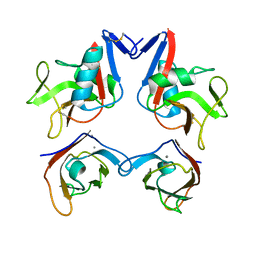 | | Structure of NK cell receptor KLRG1 bound to E-cadherin | | Descriptor: | CALCIUM ION, Epithelial cadherin, Killer cell lectin-like receptor subfamily G member 1 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
2QM4
 
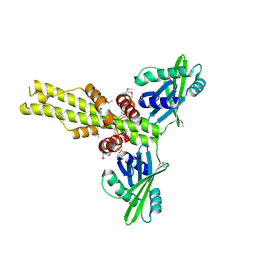 | | Crystal structure of human XLF/Cernunnos, a non-homologous end-joining factor | | Descriptor: | Non-homologous end-joining factor 1 | | Authors: | Li, Y, Chirgadze, D.Y, Sibanda, B.L, Bolanos-Garcia, V.M, Davies, O.R, Blundell, T.L. | | Deposit date: | 2007-07-14 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human XLF/Cernunnos reveals unexpected differences from XRCC4 with implications for NHEJ.
Embo J., 27, 2008
|
|
1WPA
 
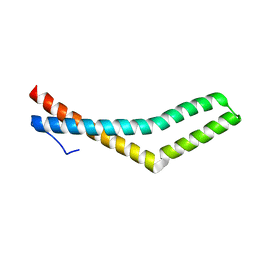 | |
5ZN9
 
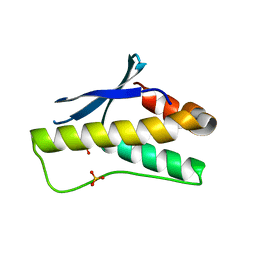 | | Crystal structure of PX domain | | Descriptor: | SULFATE ION, Sorting nexin-27 | | Authors: | Li, Y, Zhu, Z, Li, F, Liao, S, Xu, C. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Crystal structure of PX domain
To Be Published
|
|
8W7J
 
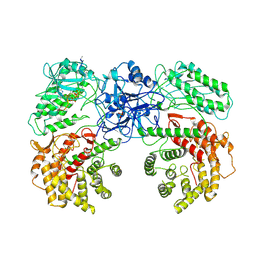 | | Cryo-EM structure of ClassIII Lanthipeptide modification enzyme PneKC with chain A bounded to substrate PneA and GTP. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, PneA LP, ... | | Authors: | Li, Y, Luo, M, Shao, K, Li, J, Li, Z. | | Deposit date: | 2023-08-30 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers.
Nat Commun, 15, 2024
|
|
8W8F
 
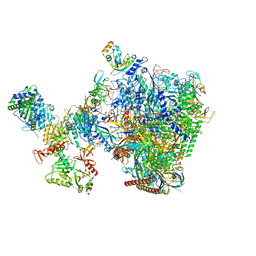 | | human co-transcriptional RNA capping enzyme RNGTT-CMTR1 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (36-MER), DNA (45-MER), ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
8W8E
 
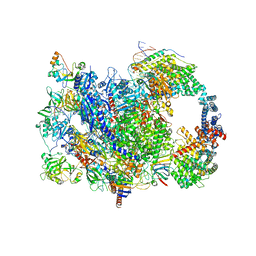 | | human co-transcriptional RNA capping enzyme RNGTT | | Descriptor: | DNA (36-MER), DNA (45-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
1YP0
 
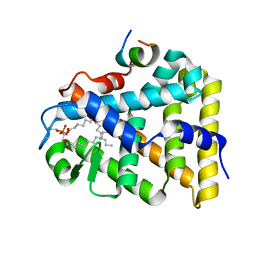 | | Structure of the steroidogenic factor-1 ligand binding domain bound to phospholipid and a SHP peptide motif | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor subfamily 0, group B, ... | | Authors: | Li, Y, Choi, M, Cavey, G, Daugherty, J, Suino, K, Kovach, A, Bingham, N, Kliewer, S, Xu, H. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1.
Mol.Cell, 17, 2005
|
|
3IBP
 
 | | The Crystal Structure of the Dimerization Domain of Escherichia coli Structural Maintenance of Chromosomes Protein MukB | | Descriptor: | AMMONIUM ION, Chromosome partition protein mukB | | Authors: | Li, Y, Schoeffler, A.J, Berger, J.M, Oakley, M.G. | | Deposit date: | 2009-07-16 | | Release date: | 2010-01-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | The crystal structure of the hinge domain of the Escherichia coli structural maintenance of chromosomes protein MukB.
J.Mol.Biol., 395, 2010
|
|
3IWP
 
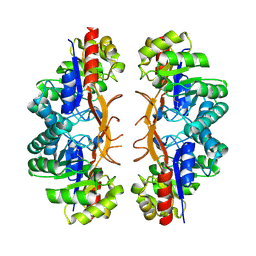 | |
8W7A
 
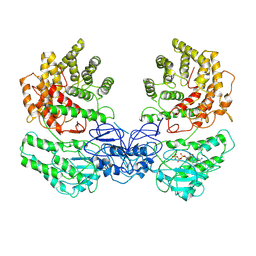 | | Cryo-EM structure of ClassIII Lanthipeptide modification enzyme PneKC in the presence of GTP. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Protein kinase domain-containing protein | | Authors: | Li, Y, Luo, M, Shao, K, Li, J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers.
Nat Commun, 15, 2024
|
|
8WGO
 
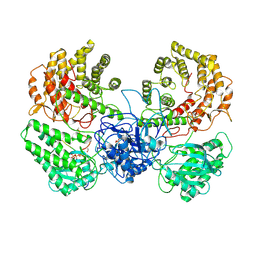 | | Cryo-EM structure of ClassIII Lanthipeptide modification enzyme PneKC in the presence of PneA and GTPrS. | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Li, Y, Luo, M, Shao, K, Li, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers.
Nat Commun, 15, 2024
|
|
3M5D
 
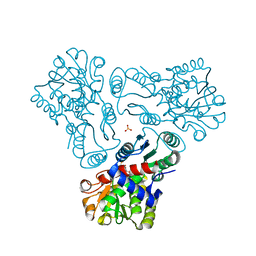 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302R mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
8JOL
 
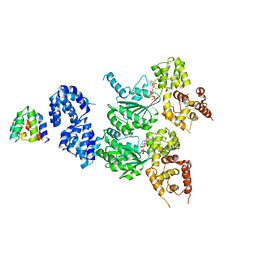 | | cryo-EM structure of the CED-4/CED-3 holoenzyme | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell death protein 3, Cell death protein 4, ... | | Authors: | Li, Y, Tian, L, Zhang, Y, Shi, Y. | | Deposit date: | 2023-06-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into CED-3 activation.
Life Sci Alliance, 6, 2023
|
|
1XOO
 
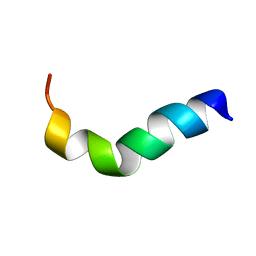 | | NMR structure of G1S mutant of influenza hemagglutinin fusion peptide in DPC micelles at pH 5 | | Descriptor: | Hemagglutinin | | Authors: | Li, Y, Han, X, Lai, A.L, Bushweller, J.H, Cafiso, D.S, Tamm, L.K. | | Deposit date: | 2004-10-06 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion.
J.Virol., 79, 2005
|
|
1ZGY
 
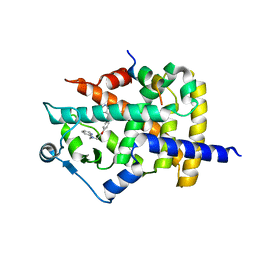 | | Structural and Biochemical Basis for Selective Repression of the Orphan Nuclear Receptor LRH-1 by SHP | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), Nuclear receptor subfamily 0, ... | | Authors: | Li, Y, Choi, M, Suino, K, Kovach, A, Daugherty, J, Kliewer, S.A, Xu, H.E. | | Deposit date: | 2005-04-22 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZH7
 
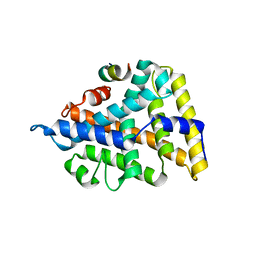 | | Structural and Biochemical Basis for Selective Repression of the Orphan Nuclear Receptor LRH-1 by SHP | | Descriptor: | Orphan nuclear receptor NR5A2, nuclear receptor subfamily 0, group B, ... | | Authors: | Li, Y, Choi, M, Suino, K, Kovach, A, Daugherty, J, Kliewer, S.A, Xu, H.E. | | Deposit date: | 2005-04-22 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3M4J
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-11 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reversible Post-Translational Carboxylation Modulates the Enzymatic Activity of N-Acetyl-l-ornithine Transcarbamylase.
Biochemistry, 49, 2010
|
|
3M4N
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302A mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-11 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
3M5C
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302E mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
