7N2F
 
 | | MicroED structure of human CPEB3 segment (154-161) straight polymorph phased by ARCIMBOLDO-BORGES | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2H
 
 | | X-Ray structure of a sequence variant of a repeat segment of the yeast prion New1p | | Descriptor: | prion New1p | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.102 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2I
 
 | | MicroED structure of human LECT2 (45-53) phased by ARCIMBOLDO-BORGES | | Descriptor: | LECT2 | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.402 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
4O9P
 
 | | Crystal structure of Thermus thermophilis transhydrogeanse domain II dimer SeMet derivative | | Descriptor: | NAD(P) transhydrogenase subunit alpha 2, NAD(P) transhydrogenase subunit beta | | Authors: | Leung, J.H, Yamaguchi, M, Moeller, A, Schurig-Briccio, L.A, Gennis, R.B, Potter, C.S, Carragher, B, Stout, C.D. | | Deposit date: | 2014-01-02 | | Release date: | 2014-06-11 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural biology. Division of labor in transhydrogenase by alternating proton translocation and hydride transfer.
Science, 347, 2015
|
|
7N47
 
 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988514 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-06-03 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7N2D
 
 | | MicroED structure of human zinc finger protein 292 segment (534-542) phased by ARCIMBOLDO-BORGES | | Descriptor: | DIMETHYL SULFOXIDE, zinc finger protein 292 | | Authors: | Richards, L.S, Flores, M.D, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.503 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N2E
 
 | | MicroED structure of human CPEB3 segment (154-161) straight polymorph | | Descriptor: | CPEB3 | | Authors: | Flores, M.D, Richards, L.S, Zee, C.T, Glynn, C, Gallagher-Jones, M, Sawaya, M.R. | | Deposit date: | 2021-05-29 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1 Å) | | Cite: | Fragment-Based Ab Initio Phasing of Peptidic Nanocrystals by MicroED.
Acs Bio Med Chem Au, 3, 2023
|
|
7N55
 
 | | The crystal structure of the mutant I38T PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988514 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7N8F
 
 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988288 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-06-14 | | Release date: | 2022-06-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7N8T
 
 | | Crystal Structure of AMP-bound Human JNK2 | | Descriptor: | ADENOSINE MONOPHOSPHATE, HEXAETHYLENE GLYCOL, Mitogen-activated protein kinase 9 | | Authors: | Li, L, Gurbani, D, Westover, K.D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of a Covalent Inhibitor of c-Jun N-Terminal Protein Kinase (JNK) 2/3 with Selectivity over JNK1.
J.Med.Chem., 66, 2023
|
|
3R08
 
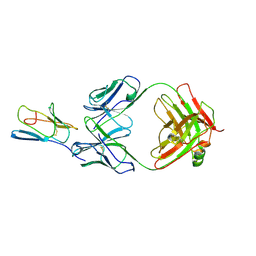 | | Crystal structure of mouse cd3epsilon in complex with antibody 2C11 Fab | | Descriptor: | Mouse anti-mouse CD3epsilon antibody 2C11 heavy chain, Mouse anti-mouse CD3epsilon antibody 2C11 light chain, T-cell surface glycoprotein CD3 epsilon chain | | Authors: | Shore, D.A, Zhu, X, Wilson, I.A. | | Deposit date: | 2011-03-07 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | T cell receptors are structures capable of initiating signaling in the absence of large conformational rearrangements.
J.Biol.Chem., 287, 2012
|
|
3R6L
 
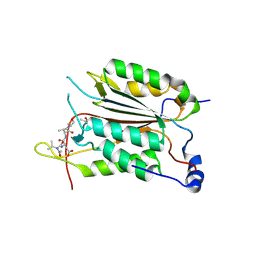 | | Caspase-2 T380A bound with Ac-VDVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)VDVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-21 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
3R7B
 
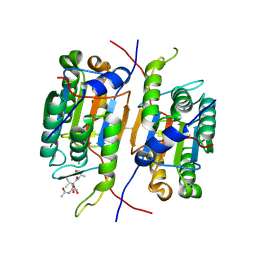 | | Caspase-2 bound to one copy of Ac-DVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)DVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
4L3Q
 
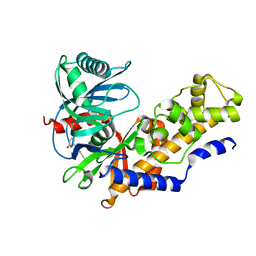 | | Crystal structure of glucokinase-activator complex | | Descriptor: | 6-{3-[(1-methyl-1H-imidazol-2-yl)sulfanyl]phenyl}pyridin-2(1H)-one, Glucokinase, alpha-D-glucopyranose | | Authors: | Gajiwala, K.S, Filipski, K.J. | | Deposit date: | 2013-06-06 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pyrimidone-based series of glucokinase activators with alternative donor-acceptor motif.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3RKX
 
 | |
4L4Y
 
 | |
7NSE
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE, H4B-FREE, ADMA COMPLEX | | Descriptor: | ACETATE ION, CACODYLIC ACID, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-01-13 | | Release date: | 2002-05-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of the Heme Domain of Bovine Endothelial Nitric Oxide Synthase Complexed with Arginine Analogues
To be Published
|
|
4LTG
 
 | | Dehydration/Rehydration of a Nucleic Acid system containing a Polypyridyl Ruthenium Complex at 74% relative humidity (2/7) | | Descriptor: | BARIUM ION, CHLORIDE ION, DNA, ... | | Authors: | Hall, J.P, Sanchez-Weatherby, J, Cardin, C.J. | | Deposit date: | 2013-07-23 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Controlled Dehydration of a Ruthenium Complex-DNA Crystal Induces Reversible DNA Kinking.
J.Am.Chem.Soc., 136, 2014
|
|
3RN2
 
 | | Structural Basis of Cytosolic DNA Recognition by Innate Immune Receptors | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*CP*AP*AP*AP*GP*AP*TP*CP*TP*TP*TP*GP*AP*TP*GP*G)-3'), Interferon-inducible protein AIM2 | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
3RIW
 
 | | The Crystal Structure of Leishmania major Peroxidase mutant C197T | | Descriptor: | Ascorbate peroxidase, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Jasion, V.S, Li, H, Poulos, T.L. | | Deposit date: | 2011-04-14 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal Structure of Leishmania major Peroxidase and Characterization of the Compound I Tryptophan Radical.
J.Biol.Chem., 286, 2011
|
|
4M5S
 
 | | Human alphaB crystallin core domain in complex with C-terminal peptide | | Descriptor: | Alpha-crystallin B chain, SUCCINIC ACID | | Authors: | Laganowsky, A, Cascio, D, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The structured core domain of alpha B-crystallin can prevent amyloid fibrillation and associated toxicity.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3U6B
 
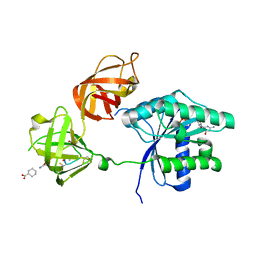 | | Ef-tu (escherichia coli) in complex with nvp-ldi028 | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Palestrant, D.J. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Antibacterial optimization of 4-aminothiazolyl analogues of the natural product GE2270 A: identification of the cycloalkylcarboxylic acids.
J.Med.Chem., 54, 2011
|
|
4OPY
 
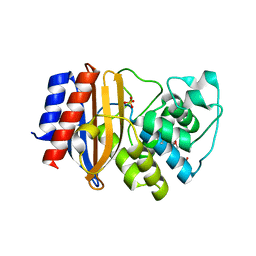 | | Room temperature crystal structure of stabilized TEM-1 beta-lactamase variant v.13 carrying R164S mutation | | Descriptor: | CALCIUM ION, Extended spectrum beta-lactamase TEM-63, SULFATE ION, ... | | Authors: | Dellus-Gur, E, Elias, M, Fraser, J.S, Tawfik, D.S. | | Deposit date: | 2014-02-07 | | Release date: | 2015-05-20 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Negative Epistasis and Evolvability in TEM-1 beta-Lactamase--The Thin Line between an Enzyme's Conformational Freedom and Disorder.
J. Mol. Biol., 427, 2015
|
|
3R7N
 
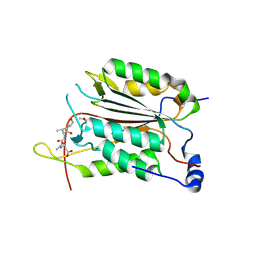 | | Caspase-2 bound with two copies of Ac-DVAD-CHO | | Descriptor: | Caspase-2 subunit p12, Caspase-2 subunit p18, Peptide Inhibitor (ACE)DVAD-CHO | | Authors: | Tang, Y, Wells, J, Arkin, M. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-27 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and enzymatic insights into caspase-2 protein substrate recognition and catalysis.
J.Biol.Chem., 286, 2011
|
|
3RLO
 
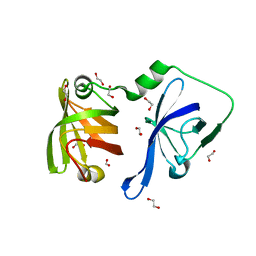 | | Structural Basis of Cytosolic DNA Recognition by Innate Receptors | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Gamma-interferon-inducible protein 16 | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
