1HC0
 
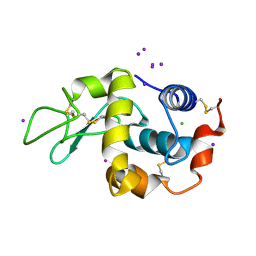 | | structure of lysozyme with periodate | | Descriptor: | CHLORIDE ION, IODIDE ION, LYSOZYME C | | Authors: | Ondracek, J, Jursik, F, Brynda, J, Rezacova, P, Sedlacek, J. | | Deposit date: | 2001-04-25 | | Release date: | 2005-11-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Regular Arrangement of Periodates Bound to Lysozyme.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2XDO
 
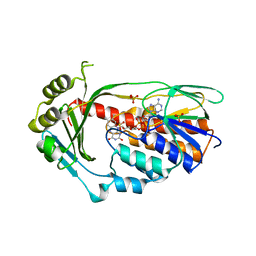 | | Structure of the Tetracycline degrading Monooxygenase TetX2 from Bacteroides thetaiotaomicron | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, TETX2 PROTEIN | | Authors: | Volkers, G, Palm, G.J, Wright, G.D, Hinrichs, W. | | Deposit date: | 2010-05-05 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Basis for a New Tetracycline Resistance Mechanism Relying on the Tetx Monooxygenase.
FEBS Lett., 585, 2011
|
|
2YIP
 
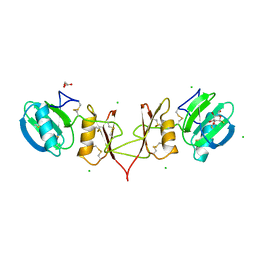 | |
2R5N
 
 | | Crystal structure of transketolase from Escherichia coli in noncovalent complex with acceptor aldose ribose 5-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 5-O-phosphono-beta-D-ribofuranose, CALCIUM ION, ... | | Authors: | Parthier, C, Asztalos, P, Wille, G, Tittmann, K. | | Deposit date: | 2007-09-04 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Strain and Near Attack Conformers in Enzymic Thiamin Catalysis: X-ray Crystallographic Snapshots of Bacterial Transketolase in Covalent Complex with Donor Ketoses Xylulose 5-phosphate and Fructose 6-phosphate, and in Noncovalent Complex with Acceptor Aldose Ribose 5-phosphate.
Biochemistry, 46, 2007
|
|
2YIO
 
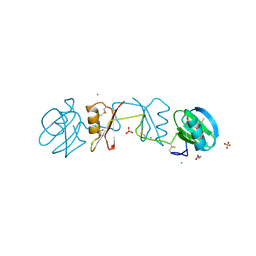 | |
2QTY
 
 | |
8BM8
 
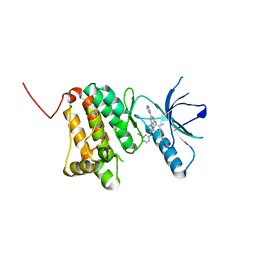 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 11 | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(3-cyanophenyl)-4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]benzamide | | Authors: | Linhard, V, Witt, K, Gande, S, Wollenhaupt, J, Lennartz, F, Weiss, M.S, Schwalbe, H. | | Deposit date: | 2022-11-10 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | (placeholder, will be filled after publication)
To Be Published
|
|
6SRF
 
 | |
6SRE
 
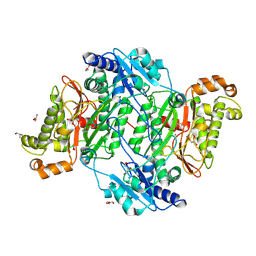 | | Crystal Structure of Human Prolidase S202F variant expressed in the presence of chaperones | | Descriptor: | GLYCEROL, GLYCINE, MANGANESE (II) ION, ... | | Authors: | Wator, E, Rutkiewicz, M, Wilk, P. | | Deposit date: | 2019-09-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Co-expression with chaperones can affect protein 3D structure as exemplified by loss-of-function variants of human prolidase.
Febs Lett., 594, 2020
|
|
4K0O
 
 | | F17b-G lectin domain with bound GlcNAc(beta1-3)Gal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-methyl beta-D-galactopyranoside, F17b-G fimbrial adhesin, NICKEL (II) ION, ... | | Authors: | Buts, L, Loris, R, Bouckaert, J, Moonens, K. | | Deposit date: | 2013-04-04 | | Release date: | 2013-04-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli
Biology (Basel), 2, 2013
|
|
6SRG
 
 | |
6TD9
 
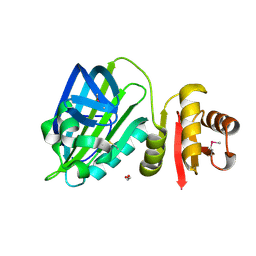 | | X-ray structure of mature PA1624 from Pseudomonas aeruginosa PAO1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PA1624, ... | | Authors: | Feiler, C.G, Blankenfeldt, W. | | Deposit date: | 2019-11-08 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The hypothetical periplasmic protein PA1624 from Pseudomonas aeruginosa folds into a unique two-domain structure.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7P27
 
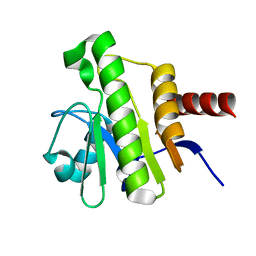 | | NMR solution structure of Chikungunya virus macro domain | | Descriptor: | Polyprotein P1234 | | Authors: | Lykouras, M.V, Tsika, A.C, Papageorgiou, N, Canard, B, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2021-07-04 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
1F1A
 
 | | Crystal structure of yeast H48Q cuznsod fals mutant analog | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1F1D
 
 | | Crystal structure of yeast H46C cuznsod mutant | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1F1G
 
 | | Crystal structure of yeast cuznsod exposed to nitric oxide | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, PHOSPHATE ION, ... | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1F18
 
 | | Crystal structure of yeast copper-zinc superoxide dismutase mutant GLY85ARG | | Descriptor: | COPPER (II) ION, COPPER-ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Hart, P.J, Ogihara, N.L, Liu, H, Nersissian, A.M, Valentine, J.S, Eisenberg, D. | | Deposit date: | 2000-05-18 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1FD9
 
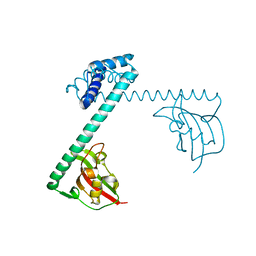 | | CRYSTAL STRUCTURE OF THE MACROPHAGE INFECTIVITY POTENTIATOR PROTEIN (MIP) A MAJOR VIRULENCE FACTOR FROM LEGIONELLA PNEUMOPHILA | | Descriptor: | PROTEIN (MACROPHAGE INFECTIVITY POTENTIATOR PROTEIN), ZINC ION | | Authors: | Riboldi-Tunnicliffe, A, Jessen, S, Konig, B, Rahfeld, J, Hacker, J, Fischer, G, Hilgenfeld, R. | | Deposit date: | 2000-07-20 | | Release date: | 2001-07-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of Mip, a prolylisomerase from Legionella pneumophila
Nat.Struct.Biol., 8, 2001
|
|
6EFG
 
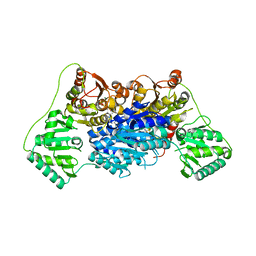 | | Pyruvate decarboxylase from Kluyveromyces lactis | | Descriptor: | MAGNESIUM ION, Pyruvate decarboxylase, THIAMINE DIPHOSPHATE | | Authors: | Kutter, S, Konig, S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structures of pyruvate decarboxylase from Kluyveromyces lactis in the absence of ligands and in the presence of the substrate surrogate pyruvamide
To be Published
|
|
6EFH
 
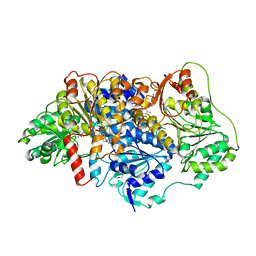 | | Pyruvate decarboxylase from Kluyveromyces lactis soaked with pyruvamide | | Descriptor: | (1S,2S)-1-amino-1,2-dihydroxypropan-1-olate, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Kutter, S, Konig, S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The crystal structures of pyruvate decarboxylase from Kluyveromyces lactis in the absence of ligands and in the presence of the substrate surrogate pyruvamide
To be Published
|
|
4Y3E
 
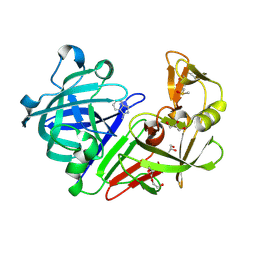 | | Endothiapepsin in complex with fragment 5 | | Descriptor: | 1H-isoindol-3-amine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y4U
 
 | | Endothiapepsin in complex with fragment 14 | | Descriptor: | 1-(4-bromo-2-chlorophenyl)-3-methylthiourea, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y3W
 
 | | Endothiapepsin in complex with fragment 35 | | Descriptor: | (2R)-2-methyl-2-(morpholin-4-yl)butan-1-amine, 1,2-ETHANEDIOL, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y3Z
 
 | | Endothiapepsin in complex with fragment 41 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y43
 
 | | Endothiapepsin in complex with fragment 42 | | Descriptor: | (1E,2E)-N,N'-di(propan-2-yl)cyclohepta-3,5-diene-1,2-diimine, 1,2-ETHANEDIOL, Endothiapepsin, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M.S, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
