7B60
 
 | | Crystal structure of MurE from E.coli in complex with Z1269139261 | | Descriptor: | 4-[(furan-2-yl)methyl]-1lambda~6~,4-thiazinane-1,1-dione, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7B6K
 
 | | Crystal structure of MurE from E.coli in complex with Z57715447 | | Descriptor: | 5-cyclohexyl-3-(pyridin-4-yl)-1,2,4-oxadiazole, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7B61
 
 | | Crystal structure of MurE from E.coli in complex with Z57299526 | | Descriptor: | (R)-N-(1-cyclopropylethyl)-6-methylpicolinamide, (S)-N-(1-cyclopropylethyl)-6-methylpicolinamide, CITRIC ACID, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7B6O
 
 | | Crystal structure of E.coli MurE mutant - C269S C340S C450S | | Descriptor: | ISOPROPYL ALCOHOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7AVQ
 
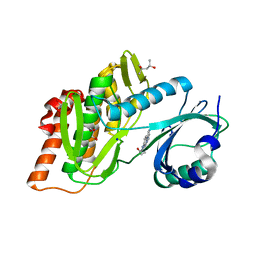 | | Crystal structure of haspin in complex with disubstituted imidazo[1,2- b]pyridazine inhibitor (compound 12) | | Descriptor: | (2~{R})-2-[[3-(2~{H}-indazol-5-yl)imidazo[1,2-b]pyridazin-6-yl]amino]butan-1-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, ... | | Authors: | Chaikuad, A, Bonnet, P, Routier, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design of new disubstituted imidazo[1,2- b ]pyridazine derivatives as selective Haspin inhibitors. Synthesis, binding mode and anticancer biological evaluation.
J Enzyme Inhib Med Chem, 35, 2020
|
|
7B6L
 
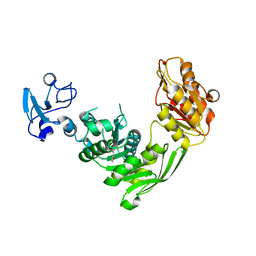 | | Crystal structure of MurE from E.coli in complex with Z57299368 | | Descriptor: | (1-ethyl-1H-benzoimidazol-2-yl)-furan-2-ylmethyl-aminee, ISOPROPYL ALCOHOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7AV8
 
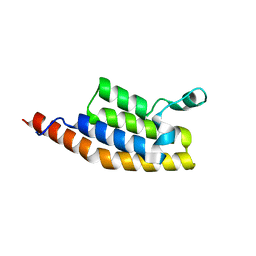 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212 | | Descriptor: | PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
To Be Published
|
|
7B36
 
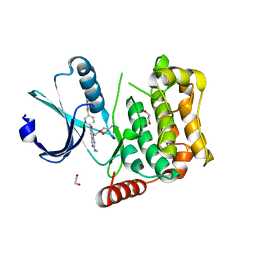 | | MST4 in complex with compound G-5555 | | Descriptor: | 1,2-ETHANEDIOL, 8-[(trans-5-amino-1,3-dioxan-2-yl)methyl]-6-[2-chloro-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7(8H)-one, CHLORIDE ION, ... | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.10681081 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7B6I
 
 | | Crystal structure of MurE from E.coli in complex with Z1373445602 | | Descriptor: | 4-(3-fluoranylpyridin-2-yl)-1-methyl-piperazin-2-one, CITRIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Talon, R, Douangamath, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.069 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7B6P
 
 | | Crystal structure of E.coli MurE - C269S C340S C450S in complex with Ellman's reagent | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
5CQ2
 
 | | Crystal Structure of tandem WW domains of ITCH in complex with TXNIP peptide | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Thioredoxin-interacting protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for the regulatory role of the PPxY motifs in the thioredoxin-interacting protein TXNIP.
Biochem.J., 473, 2016
|
|
6SHZ
 
 | | p53 cancer mutant Y220C | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, GLYCEROL, ... | | Authors: | Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-08 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: the Y220X Paradigm.
Acs Chem.Biol., 15, 2020
|
|
5E1M
 
 | | Crystal structure of NTMT1 in complex with PPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
5E2A
 
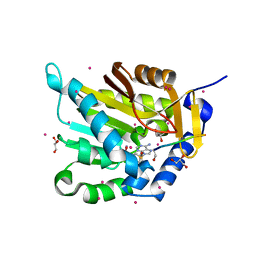 | | Crystal structure of NTMT1 in complex with N-terminally methylated SPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
4RYL
 
 | | Human Protein Arginine Methyltransferase 3 in complex with 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea | | Descriptor: | 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dobrovetsky, E, Kaniskan, H.U, Szewczyk, M, Yu, Z, Eram, M.S, Yang, X, Schmidt, K, Luo, X, Dai, M, He, F, Zang, I, Lin, Y, Kennedy, S, Li, F, Tempel, W, Smil, D, Min, S.J, Landon, M, Lin-Jones, J, Huang, X.P, Roth, B.L, Schapira, M, Atadja, P, Barsyte-Lovejoy, D, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Zhao, K, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent, Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 3 (PRMT3).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5E16
 
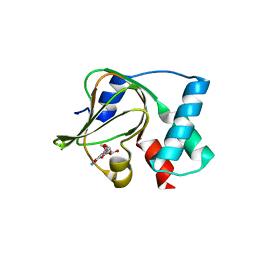 | | Co-crystal structure of the N-termial cGMP binding domain of Plasmodium falciparum PKG with cGMP | | Descriptor: | CGMP-dependent protein kinase, CYCLIC GUANOSINE MONOPHOSPHATE | | Authors: | El Bakkouri, M, Walker, J.R, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5DYL
 
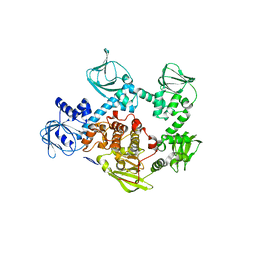 | | Crystal structure of the cGMP-dependent protein kinase PKG from Plasmodium Vivax - Apo form | | Descriptor: | cGMP-dependent protein kinase, putative | | Authors: | Wernimont, A.K, Tempel, W, He, H, Seitova, A, Hills, T, Neculai, A.M, Baker, D.A, Flueck, C, Kettleborough, C.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5DYK
 
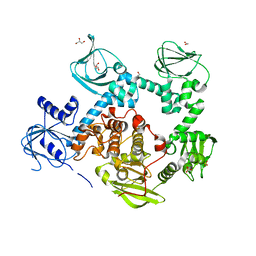 | | Crystal structure of the cGMP-dependent protein kinase PKG from Plasmodium falciparum - Apo form | | Descriptor: | 1,2-ETHANEDIOL, CGMP-dependent protein kinase, GLYCEROL, ... | | Authors: | Wernimont, A.K, Tempel, W, He, H, Seitova, A, Hills, T, Neculai, A.M, Baker, D.A, Flueck, C, Kettleborough, C.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6NFX
 
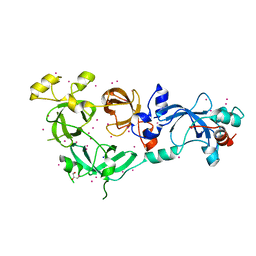 | | MBTD1 MBT repeats | | Descriptor: | GLYCEROL, MBT domain-containing protein 1,Enhancer of polycomb homolog 1, SODIUM ION, ... | | Authors: | Zhang, H, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-21 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for EPC1-Mediated Recruitment of MBTD1 into the NuA4/TIP60 Acetyltransferase Complex.
Cell Rep, 30, 2020
|
|
5E2B
 
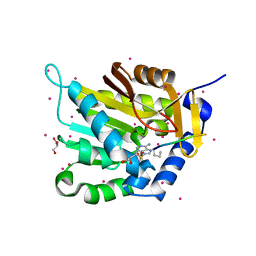 | | Crystal structure of NTMT1 in complex with N-terminally methylated PPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-30 | | Release date: | 2015-10-28 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
7B9W
 
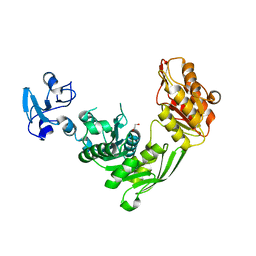 | | Crystal structure of MurE from E.coli in complex with Z757284380 | | Descriptor: | 1,2-ETHANEDIOL, 3-Chloro-N-[2-(2,4-dioxo-1,3-thiazolidin-3-yl)ethyl]benzamide, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-14 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
8P9G
 
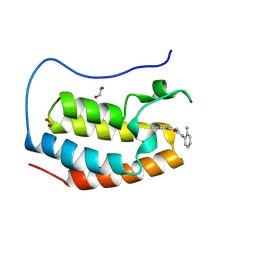 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB390 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-[(~{E})-(6-methyl-7-oxidanyl-1~{H}-indol-4-yl)diazenyl]benzamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9I
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB462 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)benzamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9K
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB503 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-azanyl-5-phenyl-phenyl)-4-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)benzamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
8P9H
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB437 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-2-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)quinoline-6-carboxamide | | Authors: | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
