1CRB
 
 | |
7QCW
 
 | |
6S03
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor I39LT379 | | Descriptor: | 4-[[4-[5,5-dimethyl-2-(6-methylpyridin-2-yl)-4,6-dihydropyrrolo[1,2-b]pyrazol-3-yl]pyridin-2-yl]amino]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
4YXX
 
 | |
6XTH
 
 | | NMR solution structure of class IV lasso peptide felipeptin A1 from Amycolatopsis sp. YIM10 | | Descriptor: | Felipeptin A1 | | Authors: | Madland, E, Guerrero-Garzon, J.F, Zotchev, S.B, Aachmann, F.L, Courtade, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Class IV Lasso Peptides Synergistically Induce Proliferation of Cancer Cells and Sensitize Them to Doxorubicin.
Iscience, 23, 2020
|
|
6XTI
 
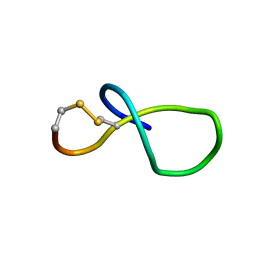 | | NMR solution structure of class IV lasso peptide felipeptin A2 from Amycolatopsis sp. YIM10 | | Descriptor: | Felipeptin A2 | | Authors: | Madland, E, Aachmann, F.L, Guerrero-Garzon, J.F, Zotchev, S.B, Courtade, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Class IV Lasso Peptides Synergistically Induce Proliferation of Cancer Cells and Sensitize Them to Doxorubicin.
Iscience, 23, 2020
|
|
6RZX
 
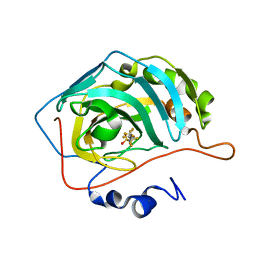 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor FBSA | | Descriptor: | 1,1,2,2,3,3,4,4,4-nonakis(fluoranyl)butane-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
6SYI
 
 | |
6V2F
 
 | | Crystal structure of the HIV capsid hexamer bound to the small molecule long-acting inhibitor, GS-6207 | | Descriptor: | HIV-1 capsid, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Appleby, T.C, Link, J.O, Yant, S.R, Villasenor, A.G, Somoza, J.R, Hu, E.Y, Schroeder, S.D, Cihlar, T. | | Deposit date: | 2019-11-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Clinical targeting of HIV capsid protein with a long-acting small molecule.
Nature, 584, 2020
|
|
5ILO
 
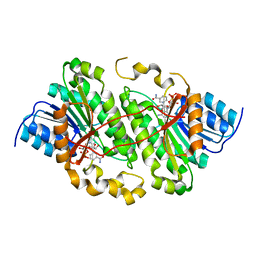 | | Crystal structure of photoreceptor dehydrogenase from Drosophila melanogaster | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Photoreceptor dehydrogenase, isoform C | | Authors: | Hofmann, L, Tsybovsky, Y, Banerjee, S. | | Deposit date: | 2016-03-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family.
Biochemistry, 55, 2016
|
|
5ILG
 
 | | Crystal structure of photoreceptor dehydrogenase from Drosophila melanogaster | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hofmann, L, Tsybovsky, Y, Banerjee, S. | | Deposit date: | 2016-03-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family.
Biochemistry, 55, 2016
|
|
6YP4
 
 | |
8P3E
 
 | | Crystal structure of glucocerebrosidase in complex with allosteric activator | | Descriptor: | 2-[[3-[(4-chlorophenyl)carbamoyl]phenyl]sulfonylamino]benzoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schulze, M.-S. | | Deposit date: | 2023-05-17 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of ss-Glucocerebrosidase Activators for Glucosylceramide hydrolysis.
Chemmedchem, 19, 2024
|
|
8P41
 
 | | Crystal structure of glucocerebrosidase in complex with allosteric activator | | Descriptor: | 2-[2-[[3,5-bis(trifluoromethyl)phenyl]methylsulfanyl]ethanoylamino]-5-chloranyl-benzoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schulze, M.-S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Identification of ss-Glucocerebrosidase Activators for Glucosylceramide hydrolysis.
Chemmedchem, 19, 2024
|
|
1TKX
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW490745 | | Descriptor: | 4-[(CYCLOPROPYLETHYNYL)OXY]-6-FLUORO-3-ISOPROPYLQUINOLIN-2(1H)-ONE, Pol polyprotein, Reverse transcriptase, ... | | Authors: | Ren, J, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 2.
J.Med.Chem., 47, 2004
|
|
8FXQ
 
 | | The Crystal Sturucture of Rhizopuspepsin with a bound modified peptide inhibitor generated by de novo drug design. | | Descriptor: | ALA-CYS-VAL-LYS, CYCLOHEXANE, Rhizopuspepsin, ... | | Authors: | Satyshur, K.A, Rich, D.H, Ripka, A.S. | | Deposit date: | 2023-01-25 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Aspartic protease inhibitors designed from computer-generated templates bind as predicted.
Org Lett, 3, 2001
|
|
6RSR
 
 | | TBK1 in complex with compound 2 | | Descriptor: | Serine/threonine-protein kinase TBK1, ~{N}-(cyclopropen-1-ylmethyl)-2-[[4-[[4-[3,3,3-tris(fluoranyl)propanoyl]piperazin-1-yl]methyl]pyridin-2-yl]amino]-1~{H}-benzimidazole-5-carboxamide | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
1CFB
 
 | | CRYSTAL STRUCTURE OF TANDEM TYPE III FIBRONECTIN DOMAINS FROM DROSOPHILA NEUROGLIAN AT 2.0 ANGSTROMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DROSOPHILA NEUROGLIAN, ... | | Authors: | Huber, A.H, Wang, Y.E, Bieber, A.J, Bjorkman, P.J. | | Deposit date: | 1994-08-27 | | Release date: | 1994-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of tandem type III fibronectin domains from Drosophila neuroglian at 2.0 A.
Neuron, 12, 1994
|
|
6RST
 
 | | TBK1 in complex with inhibitor compound 24 | | Descriptor: | 1-[4-[(1~{R})-1-[2-[[5-[1-(cyclopropylmethyl)pyrazol-4-yl]-1~{H}-benzimidazol-2-yl]amino]pyridin-4-yl]ethyl]piperazin-1-yl]-3,3,3-tris(fluoranyl)propan-1-one, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
8ATZ
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with SA112 (compound 2). | | Descriptor: | 2-[4-chloranyl-6-[[3-(2-phenylethoxy)phenyl]amino]pyrimidin-2-yl]sulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Arifi, S, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-08-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
5CVI
 
 | | Structure of the manganese regulator SloR | | Descriptor: | 1,2-ETHANEDIOL, SloR, ZINC ION | | Authors: | Nye, D, Glasfeld, A. | | Deposit date: | 2015-07-27 | | Release date: | 2015-08-12 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | SloR structure and DNA binding studies inform the SloR-SRE interaction in Streptococcus mutans
to be published
|
|
4YXY
 
 | |
4YYE
 
 | |
4ZG9
 
 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(11aS)-6-(4-fluorobenzyl)-1,3-dioxo-5,6,11,11a-tetrahydro-1H-imidazo[1',5':1,6]pyrido[3,4-b]indol-2(3H)-yl]propanoic acid, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
6SUZ
 
 | |
