2RFL
 
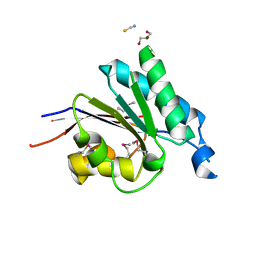 | | Crystal structure of the putative phosphohistidine phosphatase SixA from Agrobacterium tumefaciens | | Descriptor: | ACETIC ACID, GLYCEROL, Putative phosphohistidine phosphatase SixA, ... | | Authors: | Kim, Y, Binkowski, T, Xu, X, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-01 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Putative Phosphohistidine Phosphatase SixA from
Agrobacterium tumefaciens.
To be Published
|
|
1MKZ
 
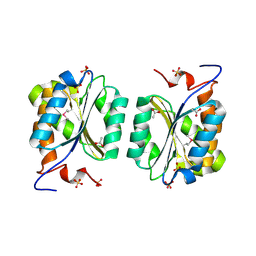 | | Crystal structure of MoaB protein at 1.6 A resolution. | | Descriptor: | ACETIC ACID, Molybdenum cofactor biosynthesis protein B, SULFATE ION | | Authors: | Sanishvili, R, Skarina, T, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-29 | | Release date: | 2003-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Escherichia coli MoaB suggests a probable role in molybdenum cofactor synthesis.
J.Biol.Chem., 279, 2004
|
|
6NKC
 
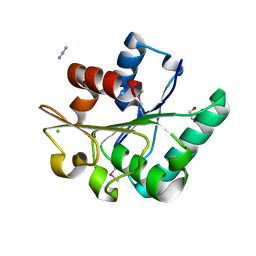 | | Crystal Structure of the Lipase Lip_vut1 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, CHLORIDE ION, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut1 from Goat Rumen metagenome.
To Be Published
|
|
2R48
 
 | | Crystal structure of the fructose specific IIB subunit of PTS system from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | Phosphotransferase system (PTS) mannose-specific enzyme IIBCA component | | Authors: | Nocek, B, Cuff, M, Sather, A, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-08-30 | | Release date: | 2007-09-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the fructose specific IIB subunit of PTS system from Bacillus subtilis subsp. subtilis str. 168.
To be Published
|
|
2RFQ
 
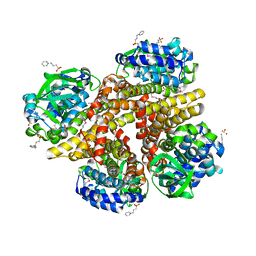 | | Crystal structure of 3-HSA hydroxylase from Rhodococcus sp. RHA1 | | Descriptor: | 3-HSA hydroxylase, oxygenase, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE | | Authors: | Chang, C, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-01 | | Release date: | 2007-10-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of 3-HSA hydroxylase, oxygenase from Rhodococcus sp. RHA1.
To be Published
|
|
2P7J
 
 | |
6NKF
 
 | | Crystal Structure of the Lipase Lip_vut4 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, 2-BUTANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.232 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut4 from Goat Rumen metagenome.
To Be Published
|
|
5DYF
 
 | | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid | | Descriptor: | Aminopeptidase N, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nocek, B, Joachimiak, A, Vassiliou, S, Berlicki, L, Mucha, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid
To Be Published
|
|
1NQK
 
 | | Structural Genomics, Crystal structure of Alkanesulfonate monooxygenase | | Descriptor: | Alkanesulfonate monooxygenase | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-21 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the protein
Alkanesulfonate monooxygenase from E. Coli
To be Published
|
|
2PQQ
 
 | | Structural Genomics, the crystal structure of the N-terminal domain of a transcriptional regulator from Streptomyces coelicolor A3(2) | | Descriptor: | FORMIC ACID, Putative transcriptional regulator | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-02 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the N-terminal domain of a transcriptional regulator from Streptomyces coelicolor A3(2)
To be Published
|
|
6NKD
 
 | | Crystal Structure of the Lipase Lip_vut3 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut3 from Goat Rumen metagenome.
To Be Published
|
|
5ES2
 
 | | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-16 | | Release date: | 2015-12-16 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To Be Published
|
|
1NR9
 
 | | Crystal Structure of Escherichia coli 1262 (APC5008), Putative Isomerase | | Descriptor: | MAGNESIUM ION, Protein YCGM | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-24 | | Release date: | 2003-07-29 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Escherichia coli Putative Isomerase EC1262 (APC5008)
To be Published
|
|
5DS0
 
 | | Crystal structure of TET aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon SCGC AB-539-E09 | | Descriptor: | COBALT (II) ION, GLYCEROL, Peptidase M42 | | Authors: | Michalska, K, Chhor, G, Mootz, J, Endres, M, Jedrzejczak, R, Babnigg, G, Steen, A, Lloyd, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of TET aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon SCGC AB-539-E09
To Be Published
|
|
4EW5
 
 | | C-terminal domain of inner membrane protein CigR from Salmonella enterica. | | Descriptor: | 1,2-ETHANEDIOL, CigR Protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Niemann, G.S, Merkley, E.D, Savchenko, A, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | C-terminal domain of inner membrane protein CigR from Salmonella enterica.
To be Published
|
|
4GYT
 
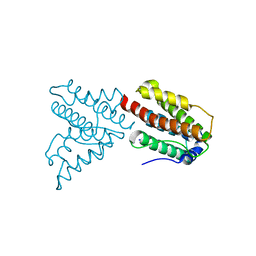 | | Crystal structure of lpg0076 protein from Legionella pneumophila | | Descriptor: | SODIUM ION, uncharacterized protein | | Authors: | Michalska, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Crystal structure of lpg0076 protein from Legionella pneumophila
To be Published
|
|
4H2K
 
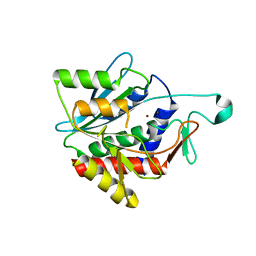 | | Crystal structure of the catalytic domain of succinyl-diaminopimelate desuccinylase from Haemophilus influenzae | | Descriptor: | Succinyl-diaminopimelate desuccinylase, ZINC ION | | Authors: | Nocek, B, Jedrzejczak, R, Makowska-Grzyska, M, Starus, A, Holz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-12 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The dimerization domain in DapE enzymes is required for catalysis.
Plos One, 9, 2014
|
|
2QM0
 
 | | Crystal structure of BES protein from Bacillus cereus | | Descriptor: | BES, SULFATE ION | | Authors: | Kim, Y, Maltseva, N, Zawadzka, A, Holzle, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-13 | | Release date: | 2007-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of BES from Bacillus cereus.
To be Published
|
|
4K0D
 
 | | Periplasmic sensor domain of sensor histidine kinase, Adeh_2942 | | Descriptor: | ACETATE ION, CHLORIDE ION, Periplasmic sensor hybrid histidine kinase, ... | | Authors: | Pokkuluri, P.R, Mack, J.C, Bearden, J, Rakowski, E, Schiffer, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-04-03 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Analysis of periplasmic sensor domains from Anaeromyxobacter dehalogenans 2CP-C: structure of one sensor domain from a histidine kinase and another from a chemotaxis protein.
Microbiologyopen, 2, 2013
|
|
2QPV
 
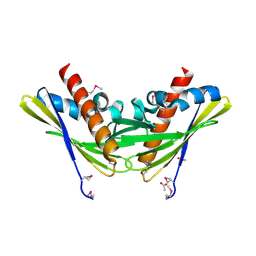 | | Crystal structure of uncharacterized protein Atu1531 | | Descriptor: | ACETIC ACID, Uncharacterized protein Atu1531 | | Authors: | Chang, C, Binkowski, T.A, Xu, X, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-25 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of uncharacterized protein Atu1531.
To be Published
|
|
2R8B
 
 | | The crystal structure of the protein Atu2452 of unknown function from Agrobacterium tumefaciens str. C58 | | Descriptor: | SULFATE ION, Uncharacterized protein Atu2452 | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-10 | | Release date: | 2007-09-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The crystal structure of the protein Atu2452 of unknown function from Agrobacterium tumefaciens str. C58.
TO BE PUBLISHED
|
|
2R78
 
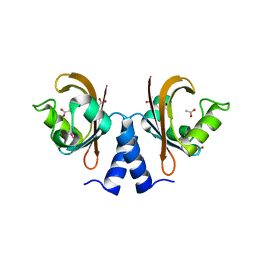 | |
4E94
 
 | | Crystal structure of MccF-like protein from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, MccC family protein, SULFATE ION | | Authors: | Nocek, B, Tikhonov, A, Kwon, K, Severinov, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal structure of MccF-like protein from Streptococcus pneumoniae
TO BE PUBLISHED
|
|
1P8C
 
 | | Crystal structure of TM1620 (APC4843) from Thermotoga maritima | | Descriptor: | conserved hypothetical protein | | Authors: | Kim, Y, Joachimiak, A, Brunzelle, J.S, Korolev, S.V, Edwards, A, Xu, X, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-05-06 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of Thermotoga maritima protein TM1620 (APC4843)
To be Published
|
|
1ORU
 
 | | Crystal Structure of APC1665, YUAD protein from Bacillus subtilis | | Descriptor: | CHLORIDE ION, SULFATE ION, yuaD protein | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-03-15 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of APC1665, YUAD protein from Bacillus subtilis
To be Published
|
|
