5VKC
 
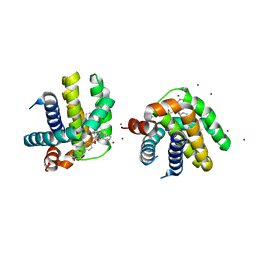 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(3-{[4-(4-acetylpiperazin-1-yl)phenoxy]methyl}-1,5-dimethyl-1H-pyrazol-4-yl)-3-{3-[(naphthalen-1-yl)oxy]propyl}-1-[(pyridin-3-yl)methyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
3DDS
 
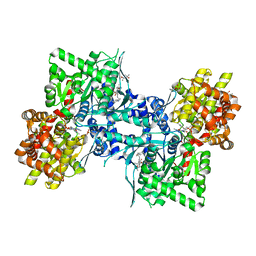 | |
5UKY
 
 | | DHp domain of PhoR of M. tuberculosis - native data | | Descriptor: | 1,2-ETHANEDIOL, ATP-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, S. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Asymmetric Structure of the Dimerization Domain of PhoR, a Sensor Kinase Important for the Virulence of Mycobacterium tuberculosis.
ACS Omega, 2, 2017
|
|
5UKV
 
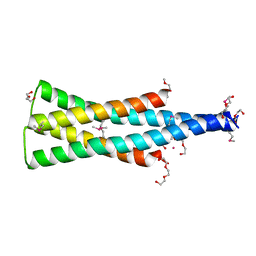 | | DHp domain of PhoR of M. tuberculosis - SeMet | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ATP-binding protein, ... | | Authors: | Wang, S. | | Deposit date: | 2017-01-23 | | Release date: | 2017-12-06 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Asymmetric Structure of the Dimerization Domain of PhoR, a Sensor Kinase Important for the Virulence of Mycobacterium tuberculosis.
ACS Omega, 2, 2017
|
|
3DDW
 
 | | Crystal structure of glycogen phosphorylase complexed with an anthranilimide based inhibitor GSK055 | | Descriptor: | (2S)-{[(3-{[(2-chloro-6-methylphenyl)carbamoyl]amino}naphthalen-2-yl)carbonyl]amino}(phenyl)ethanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2008-06-06 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Anthranilimide based glycogen phosphorylase inhibitors for the treatment of type 2 diabetes. Part 3: X-ray crystallographic characterization, core and urea optimization and in vivo efficacy.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3DD1
 
 | | Crystal structure of glycogen phophorylase complexed with an anthranilimide based inhibitor GSK254 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-cyclohexyl-N-[(3-{[(2,4,6-trimethylphenyl)carbamoyl]amino}naphthalen-2-yl)carbonyl]-D-alanine, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2008-06-04 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Anthranilimide based glycogen phosphorylase inhibitors for the treatment of type 2 diabetes. Part 3: X-ray crystallographic characterization, core and urea optimization and in vivo efficacy.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8JYM
 
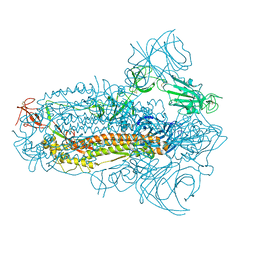 | | Structure of the SARS-CoV-2 XBB.1.5 spike glycoprotein (closed state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYK
 
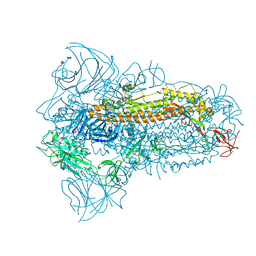 | | Structure of the SARS-CoV-2 XBB.1.5 spike glycoprotein (closed state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYP
 
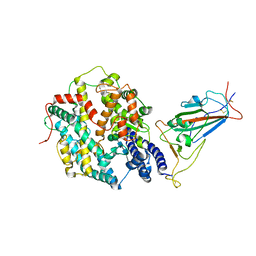 | | Structure of SARS-CoV-2 XBB.1.5 spike RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYN
 
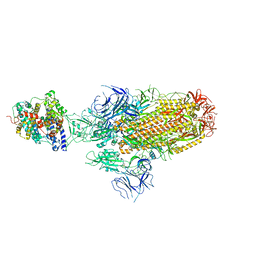 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYO
 
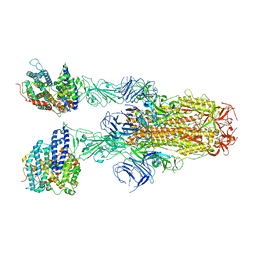 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (2-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
7CMW
 
 | | Complex structure of PARP1 catalytic domain with pamiparib | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 1 | | Authors: | Feng, Y.C, Peng, H, Hong, Y, Liu, Y. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Pamiparib (BGB-290), a Potent and Selective Poly (ADP-ribose) Polymerase (PARP) Inhibitor in Clinical Development.
J.Med.Chem., 63, 2020
|
|
8KFX
 
 | | Gi bound CCR8 complex with nonpeptide agonist LMD-009 | | Descriptor: | 8-[[3-(2-methoxyphenoxy)phenyl]methyl]-1-(2-phenylethyl)-1,3,8-triazaspiro[4.5]decan-4-one, C-C chemokine receptor type 8,Fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
8KFZ
 
 | | Gi bound CCR8 in ligand free state | | Descriptor: | C-C chemokine receptor type 8,LgBiT fusion protein,Recombinant Human Rhinovirus, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
8KFY
 
 | | Gi bound CCR8 complex with nonpeptide agonist ZK 756326 | | Descriptor: | 2-[2-[4-[(3-phenoxyphenyl)methyl]piperazin-1-yl]ethoxy]ethanol, C-C chemokine receptor type 8,Fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
6IR5
 
 | | P domain of GII.3-TV24 | | Descriptor: | VP1 Capsid protein | | Authors: | Yang, Y. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
6IS5
 
 | | P domain of GII.3-TV24 with A-tetrasaccharide complex | | Descriptor: | VP1 Capsid protein, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yang, Y. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
6IK4
 
 | | A Novel M23 Metalloprotease Pseudoalterin from Deep-sea | | Descriptor: | Elastinolytic metalloprotease, GLYCEROL, ZINC ION | | Authors: | Zhao, H.L, Tang, B.L, Yang, J, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-10-14 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A predator-prey interaction between a marine Pseudoalteromonas sp. and Gram-positive bacteria.
Nat Commun, 11, 2020
|
|
3CID
 
 | | Structure of BACE Bound to SCH726222 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2S)-2-[(4S)-1-benzyl-5-oxoimidazolidin-4-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CIB
 
 | | Structure of BACE Bound to SCH727596 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N'-[(1S,2R)-2-[(2R,4S)-4-benzylpiperidin-2-yl]-1-(3,5-difluorobenzyl)-2-hydroxyethyl]-5-methyl-N,N-dipropylbenzene-1,3-dicarboxamide | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2008-03-11 | | Release date: | 2008-06-10 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Rational design of novel, potent piperazinone and imidazolidinone BACE1 inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6VIZ
 
 | | BRD4_Bromodomain1 complex with pyrrolopyridone compound 27 | | Descriptor: | 4-[2-(2,6-dimethylphenoxy)-5-(ethylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
6P7Z
 
 | |
6VIY
 
 | | BRD2_Bromodomain2 complex with pyrrolopyridone compound 27 | | Descriptor: | 4-[2-(2,6-dimethylphenoxy)-5-(ethylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 2 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
6J0Q
 
 | | Crystal structure of P domain from GII.11 swine norovirus | | Descriptor: | VP1 capsid protein | | Authors: | Chen, Y. | | Deposit date: | 2018-12-25 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of host ligand specificity change of GII porcine noroviruses from their closely related GII human noroviruses.
Emerg Microbes Infect, 8, 2019
|
|
6VIX
 
 | | BRD4_Bromodomain2 complex with pyrrolopyridone compound 18 | | Descriptor: | 4-[2-(2,4-difluorophenoxy)-5-(methylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
