5KDT
 
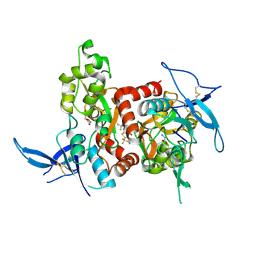 | | Structure of the human GluN1/GluN2A LBD in complex with GNE0723 | | Descriptor: | (1~{R},2~{R})-2-[7-[[5-chloranyl-3-(trifluoromethyl)pyrazol-1-yl]methyl]-5-oxidanylidene-2-(trifluoromethyl)-[1,3]thiazolo[3,2-a]pyrimidin-3-yl]cyclopropane-1-carbonitrile, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-06-08 | | Release date: | 2016-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J. Med. Chem., 59, 2016
|
|
8ZB6
 
 | |
8ZH6
 
 | |
7BZ5
 
 | | Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of B38, Light chain of B38, ... | | Authors: | Wu, Y, Qi, J, Gao, F. | | Deposit date: | 2020-04-26 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Science, 368, 2020
|
|
7EQ2
 
 | | Crystal structure of GDP-bound Rab1a-T75D | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Cao, Y.L, Gu, D.D, Gao, S. | | Deposit date: | 2021-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55090284 Å) | | Cite: | Aurora kinase A-mediated phosphorylation triggers structural alteration of Rab1A to enhance ER complexity during mitosis
Nat.Struct.Mol.Biol., 2024
|
|
6RYB
 
 | |
1J4J
 
 | | Crystal Structure of Tabtoxin Resistance Protein (form II) complexed with an Acyl Coenzyme A | | Descriptor: | ACETYL COENZYME *A, TABTOXIN RESISTANCE PROTEIN | | Authors: | He, H, Ding, Y, Bartlam, M, Zhang, R, Duke, N, Joachimiak, A, Shao, Y, Cao, Z, Tang, H, Liu, Y, Jiang, F, Liu, J, Zhao, N, Rao, Z. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of tabtoxin resistance protein complexed with acetyl coenzyme A reveals the mechanism for beta-lactam acetylation.
J.Mol.Biol., 325, 2003
|
|
6RYA
 
 | | Structure of Dup1 mutant H67A:Ubiquitin complex | | Descriptor: | Polyubiquitin-C, Septation initiation protein | | Authors: | Donghyuk, S, Ivan, D. | | Deposit date: | 2019-06-10 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Regulation of Phosphoribosyl-Linked Serine Ubiquitination by Deubiquitinases DupA and DupB.
Mol.Cell, 77, 2020
|
|
7O8C
 
 | | Structure of SGBP BO2743 from Bacteroides ovatus | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Correia, V.C, Trovao, F, Pinheiro, B.A, Palma, A.S, Carvalho, A.L. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Molecular Recognition of beta 1,3-1,4-Glucans by a Surface Glycan-Binding Protein from the Human Gut Symbiont Bacteroides ovatus.
Microbiol Spectr, 9, 2021
|
|
7O5Y
 
 | |
7OA8
 
 | |
7OA7
 
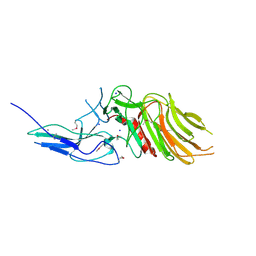 | | PilC minor pilin of Streptococcus sanguinis 2908 type IV pili | | Descriptor: | CALCIUM ION, PilC minor pilin, SODIUM ION | | Authors: | Sheppard, D, Pelicic, V. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2023-01-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of a glycan-binding complex of minor pilins completes the analysis of Streptococcus sanguinis type 4 pili subunits.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7P1C
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin B | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein assembly factor BamA, TRP-ASN-UX8-THR-LYS-ARG-PHE | | Authors: | Jakob, R.P, Modaresi, S.M, Hiller, S, Maier, T. | | Deposit date: | 2021-07-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutasynthetic Production and Antimicrobial Characterization of Darobactin Analogs.
Microbiol Spectr, 9, 2021
|
|
6O8C
 
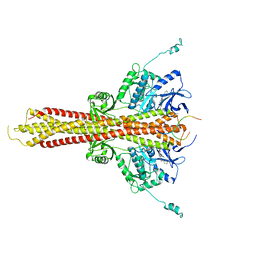 | | Crystal structure of STING CTT in complex with TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Li, P, Zhao, B, Du, F. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1.
Nature, 569, 2019
|
|
6O8B
 
 | | Crystal structure of STING CTD in complex with TBK1 | | Descriptor: | N-(3-{[5-iodo-4-({3-[(thiophen-2-ylcarbonyl)amino]propyl}amino)pyrimidin-2-yl]amino}phenyl)pyrrolidine-1-carboxamide, Serine/threonine-protein kinase TBK1, Stimulator of interferon genes protein | | Authors: | Li, P, Zhao, B, Du, F. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1.
Nature, 569, 2019
|
|
6KUR
 
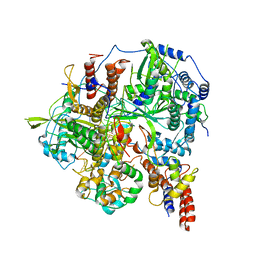 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B1) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUK
 
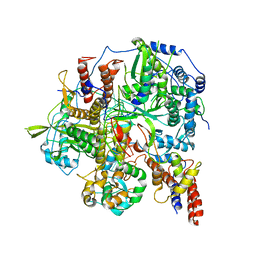 | | Structure of influenza D virus polymerase bound to vRNA promoter in mode A conformation (class A1) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUP
 
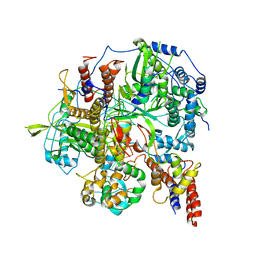 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode A conformation(Class A2) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUV
 
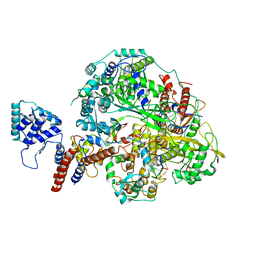 | | Structure of influenza D virus polymerase bound to cRNA promoter in class 2 | | Descriptor: | 3'-cRNA, 5'-cRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUT
 
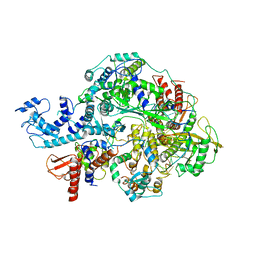 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B2) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KV5
 
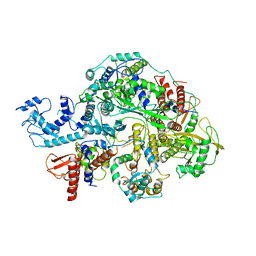 | | Structure of influenza D virus apo polymerase | | Descriptor: | Polymerase 3, Polymerase PB2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6G0C
 
 | | Crystal structure of SdeA catalytic core | | Descriptor: | 1,2-ETHANEDIOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Ubiquitinating/deubiquitinating enzyme SdeA | | Authors: | Kalayil, S, Bhogaraju, S, Basquin, J, Dikic, I. | | Deposit date: | 2018-03-17 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination.
Nature, 557, 2018
|
|
6G47
 
 | | Crystal Structure of Human Adenovirus 52 Short Fiber Knob in Complex with alpha-(2,8)-Trisialic Acid (DP3) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Liaci, A.M, Stehle, T. | | Deposit date: | 2018-03-26 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Polysialic acid is a cellular receptor for human adenovirus 52.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7BXU
 
 | | CLC-7/Ostm1 membrane protein complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H(+)/Cl(-) exchange transporter 7, Osteopetrosis-associated transmembrane protein 1 | | Authors: | Zhang, S.S, Yang, M.J. | | Deposit date: | 2020-04-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular insights into the human CLC-7/Ostm1 transporter.
Sci Adv, 6, 2020
|
|
5WWU
 
 | | Crystal Structure of HLA-A*2402 in complex with 2009 pandemic influenza A(H1N1) virus and avian influenza A(H5N1) virus-derived peptide H1-25 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-05 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
