5IV1
 
 | | Solution Structure of DNA Dodecamer with 8-oxoguanine at 4th Position | | Descriptor: | DNA (5'-D(*CP*GP*CP*(8OG)P*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Miears, H.L, Gruber, D.R, Hoppins, J.J, Kiryutin, A.S, Kasymov, R.D, Yurkovskaya, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 8-Oxoguanine Affects DNA Backbone Conformation in the EcoRI Recognition Site and Inhibits Its Cleavage by the Enzyme.
Plos One, 11, 2016
|
|
5ITP
 
 | |
5IWD
 
 | | HCMV DNA polymerase subunit UL44 complex with a small molecule | | Descriptor: | 5-methylidene-3-(methylsulfanyl)-2-benzothiophen-4(5H)-one, DNA polymerase processivity factor | | Authors: | Chen, H, Coen, D.M, Hogle, J.M, Filman, D.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A Small Covalent Allosteric Inhibitor of Human Cytomegalovirus DNA Polymerase Subunit Interactions.
ACS Infect Dis, 3, 2017
|
|
2MS0
 
 | |
5J7Q
 
 | | Macrophage Migration Inhibitory Factor bound to Inhibitor K664 Derivative | | Descriptor: | 4-(imidazo[1,2-a]pyridin-2-yl)benzene-1,2-diol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Robertson, M.J, Jorgensen, W.L. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Irregularities in enzyme assays: The case of macrophage migration inhibitory factor.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5J7T
 
 | |
5IU5
 
 | | Lambda-[Ru(TAP)2(dppz)]2+ bound to d(TCGGCICCGA)2 | | Descriptor: | BARIUM ION, DNA (5'-D(*TP*CP*GP*GP*CP*IP*CP*CP*GP*A)-3'), Ru(tap)2(dppz) complex | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2016-03-17 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unexpected enhancement of sensitised photo-oxidation by a Ru(II) complex on replacement of guanine with inosine in DNA
To Be Published
|
|
5IVG
 
 | |
5J6G
 
 | |
5J7S
 
 | | Crystal structure of SM1-71 bound to TAK1-TAB1 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7/TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]phenyl}propanamide | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | Structure-guided development of covalent TAK1 inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
2N8X
 
 | | Solution structure of LptE from Pseudomonas Aerigunosa | | Descriptor: | LPS-assembly lipoprotein LptE | | Authors: | Moehle, K, Kocherla, H, Jurt, S, Robinson, J, Zerbe, O, Zerbe, K, Bacsa, B. | | Deposit date: | 2015-10-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of LptE from Pseudomonas aeruginosa.
Biochemistry, 55, 2016
|
|
5ITO
 
 | |
5IVR
 
 | |
5J7P
 
 | |
5J8I
 
 | | Crystal structure of TL11-113 bound to TAK1-TAB1 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7/TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2016-04-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Structure-guided development of covalent TAK1 inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5JEU
 
 | | del-[Ru(phen)2(dppz)]2+ bound to d(TCGGCGCCGA) with Ba2+ | | Descriptor: | BARIUM ION, CHLORIDE ION, DNA (5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3'), ... | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2016-04-19 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Delta chirality ruthenium 'light-switch' complexes can bind in the minor groove of DNA with five different binding modes.
Nucleic Acids Res., 44, 2016
|
|
5JFQ
 
 | | Geranylgeranyl Pyrophosphate Synthetase from archaeon Geoglobus acetivorans | | Descriptor: | Geranylgeranyl Pyrophosphate Synthetase | | Authors: | Petrova, T, Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2016-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural characterization of geranylgeranyl pyrophosphate synthase GACE1337 from the hyperthermophilic archaeon Geoglobus acetivorans.
Extremophiles, 22, 2018
|
|
2MT7
 
 | |
5JH5
 
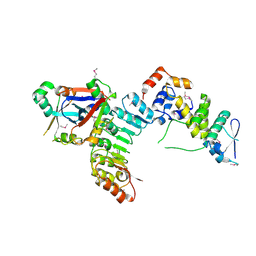 | | Structural Basis for the Hierarchical Assembly of the Core of PRC1.1 | | Descriptor: | BCL-6 corepressor-like protein 1, Lysine-specific demethylase 2B, Polycomb group RING finger protein 1, ... | | Authors: | Wong, S.J, Taylor, A.B, Hart, P.J, Kim, C.A. | | Deposit date: | 2016-04-20 | | Release date: | 2016-09-14 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | KDM2B Recruitment of the Polycomb Group Complex, PRC1.1, Requires Cooperation between PCGF1 and BCORL1.
Structure, 24, 2016
|
|
5JLJ
 
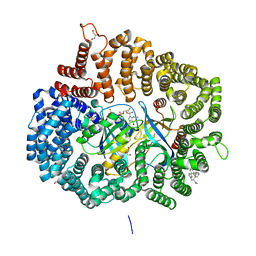 | | Crystal Structure of KPT8602 in complex with CRM1-Ran-RanBP1 | | Descriptor: | (2R)-3-{3-[3,5-bis(trifluoromethyl)phenyl]-1H-1,2,4-triazol-1-yl}-2-(pyrimidin-5-yl)propanamide, CHLORIDE ION, Exportin-1, ... | | Authors: | Fung, H.Y, Chook, Y.M. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Next-generation XPO1 inhibitor shows improved efficacy and in vivo tolerability in hematological malignancies.
Leukemia, 30, 2016
|
|
5JRS
 
 | |
5JW5
 
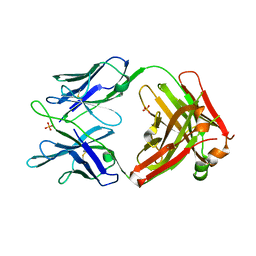 | | Structure of MEDI8852 Fab Fragment | | Descriptor: | MEDI8852 Heavy chain, MEDI8852 Light chain, PHOSPHATE ION | | Authors: | Neu, U, Collins, P.J, Walker, P.A, Vorlaender, M.K, Ogrodowicz, R.W, Martin, S.R, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2016-05-11 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes.
Cell, 166, 2016
|
|
2MS1
 
 | |
2MQV
 
 | |
2MQT
 
 | |
