2C4R
 
 | | Catalytic domain of E. coli RNase E | | Descriptor: | MAGNESIUM ION, RIBONUCLEASE E, SSRNA MOLECULE: 5'-R(*AP*CP*AP*GP*UP*AP*UP*UP*UP*GP)-3', ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Luisi, B.F. | | Deposit date: | 2005-10-21 | | Release date: | 2005-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
1O9R
 
 | | The X-ray crystal structure of Agrobacterium tumefaciens Dps, a member of the family that protect DNA without binding | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AGROBACTERIUM TUMEFACIENS DPS, ... | | Authors: | Ilari, A, Ceci, P, Chiancone, E. | | Deposit date: | 2002-12-18 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Dps Protein of Agrobacterium Tumefaciens Does not Bind to DNA But Protects It Toward Oxidative Cleavage: X-Ray Crystal Structure, Iron Binding, and Hydroxyl-Radical Scavenging Properties
J.Biol.Chem., 278, 2003
|
|
1OXY
 
 | |
4DK6
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
2F8M
 
 | |
2FVC
 
 | | Crystal structure of NS5B BK strain (delta 24) in complex with a 3-(1,1-Dioxo-2H-(1,2,4)-benzothiadiazin-3-yl)-4-hydroxy-2(1H)-quinolinone | | Descriptor: | 3-(1,1-dioxido-4H-1,2,4-benzothiadiazin-3-yl)-4-hydroxy-1-(3-methylbutyl)quinolin-2(1H)-one, polyprotein | | Authors: | Concha, N.O, Wonacott, A, Singh, O. | | Deposit date: | 2006-01-30 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-(1,1-dioxo-2H-(1,2,4)-benzothiadiazin-3-yl)-4-hydroxy-2(1H)-quinolinones, potent inhibitors of hepatitis C virus RNA-dependent RNA polymerase.
J.Med.Chem., 49, 2006
|
|
8DB4
 
 | |
7TV2
 
 | | X-ray crystal structure of HIV-2 CA protein CTD | | Descriptor: | Capsid protein p24 | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2022-02-03 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | HIV-2 Immature Particle Morphology Provides Insights into Gag Lattice Stability and Virus Maturation.
J.Mol.Biol., 435, 2023
|
|
6UFX
 
 | | WD repeat-containing protein 5 complexed with N-[(3,5-dimethoxyphenyl)methyl]-4'-fluoro-5-{[(2E)-2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl]methyl}-2'-methyl[1,1'-biphenyl]-3-carboxamide (compound 13) | | Descriptor: | N-[(3,5-dimethoxyphenyl)methyl]-4'-fluoro-5-{[(2E)-2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl]methyl}-2'-methyl[1,1'-biphenyl]-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Rietz, T.A, Fesik, S.W, Zhao, B. | | Deposit date: | 2019-09-25 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Discovery and Structure-Based Optimization of Potent and Selective WD Repeat Domain 5 (WDR5) Inhibitors Containing a Dihydroisoquinolinone Bicyclic Core.
J.Med.Chem., 63, 2020
|
|
6UCS
 
 | |
8DZK
 
 | | Dbr1 in complex with 5-mer cleavage product | | Descriptor: | FE (II) ION, RNA (5'-R(P*(G46)P*UP*GP*UP*U)-3'), RNA lariat debranching enzyme, ... | | Authors: | Clark, N.E, Taylor, A.B. | | Deposit date: | 2022-08-08 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the RNA Lariat Debranching Enzyme Dbr1 with Hydrolyzed Phosphorothioate RNA Product.
Biochemistry, 61, 2022
|
|
7UXO
 
 | | Structure of PDL1 in complex with FP30790, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, FP30790, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Travaline, T, Tattersfield, H, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UYK
 
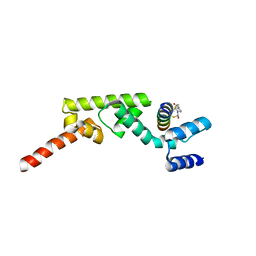 | | Structure of RNF31 in complex with FP06655, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, E3 ubiquitin-protein ligase RNF31, Helicon FP06655, ... | | Authors: | Agarwal, S, Thomson, T, Wahl, S, Walkup, W, Olsen, T, Verdine, G, McGee, J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UWO
 
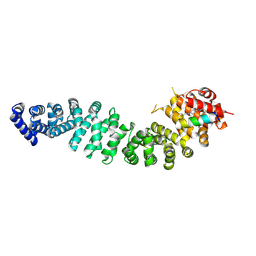 | | Structure of beta-catenin in complex with FP05874, a Helicon Polypeptide | | Descriptor: | Catenin beta-1, Helicon Polypeptide FP05874, N,N'-(1,4-phenylene)diacetamide | | Authors: | Agarwal, S, Thomson, T, Wahl, S, Ramirez, J, Hriniak, B, Verdine, G, McGee, J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXK
 
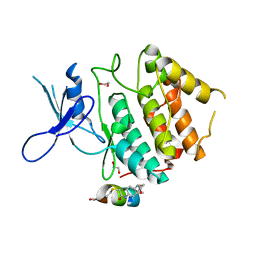 | | Structure of CDK2 in complex with FP24322, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, FP24322, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXP
 
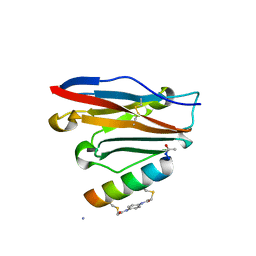 | | Structure of PDL1 in complex with FP28132, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, FP28132, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Travaline, T, Tattersfield, H, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UWI
 
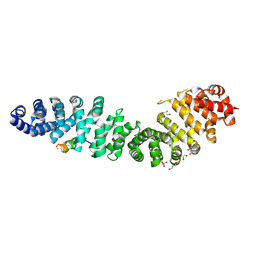 | | Structure of beta-catenin in complex with FP01567, a Helicon Polypeptide | | Descriptor: | Catenin beta-1, GLYCEROL, Helicon Polypeptide FP01567, ... | | Authors: | Brennan, M, Agarwal, S, Thomson, T, Wahl, S, Ramirez, J, Verdine, G, McGee, J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXQ
 
 | | Structure of PDL1 in complex with FP28135, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, FP28135, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Travaline, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UY2
 
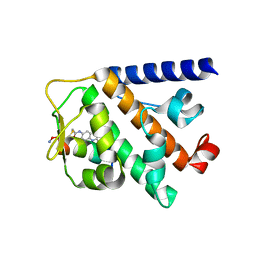 | | Structure of RNF31 in complex with FP06649, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, E3 ubiquitin-protein ligase RNF31, Helicon FP06649, ... | | Authors: | Agarwal, S, Thomson, T, Wahl, S, Walkup, W, Olsen, T, Verdine, G, McGee, J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UX5
 
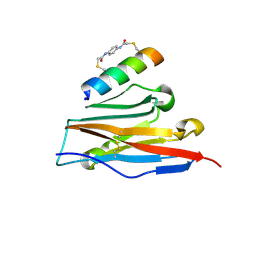 | | Structure of PDL1 in complex with FP28136, a Helicon Polypeptide | | Descriptor: | Helicon FP28136, N,N'-(1,4-phenylene)diacetamide, Programmed cell death 1 ligand 1 | | Authors: | Agarwal, S, Li, K, Tokareva, O, Thomson, T, Travaline, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXI
 
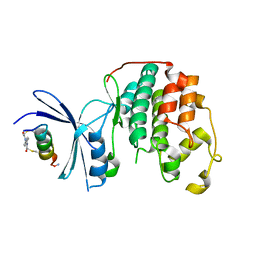 | | Structure of CDK2 in complex with FP19711, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, Cyclin-dependent kinase 2, FP19711, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXJ
 
 | | Structure of PPIA in complex with FP29102, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, FP29102, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXM
 
 | | Structure of PPIA in complex with FP29092, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, AMINO GROUP, FP29092, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UYJ
 
 | | Structure of RNF31 in complex with FP06652, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, E3 ubiquitin-protein ligase RNF31, Helicon FP06652, ... | | Authors: | Agarwal, S, Thomson, T, Wahl, S, Walkup, W, Olsen, T, Verdine, G, McGee, J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXN
 
 | | Structure of PPIA in complex with FP29103, a Helicon Polypeptide | | Descriptor: | FP29103, N,N'-(1,4-phenylene)diacetamide, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Tattersfield, H, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
