3QP4
 
 | | Crystal structure of CviR ligand-binding domain bound to C10-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QP2
 
 | | Crystal structure of CviR ligand-binding domain bound to C8-HSL | | Descriptor: | CviR transcriptional regulator, N-(2-OXOTETRAHYDROFURAN-3-YL)OCTANAMIDE | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.638 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
2POX
 
 | |
2QYP
 
 | |
5ND5
 
 | | Crystal structure of transketolase from Chlamydomonas reinhardtii in complex with TPP and Mg2+ | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, Transketolase | | Authors: | Fermani, S, Zaffagnini, M, Francia, F, Pasquini, M. | | Deposit date: | 2017-03-07 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for the magnesium-dependent activation of transketolase from Chlamydomonas reinhardtii.
Biochim. Biophys. Acta, 1861, 2017
|
|
2RB3
 
 | | Crystal Structure of Human Saposin D | | Descriptor: | GLYCEROL, Proactivator polypeptide, SULFATE ION | | Authors: | Maier, T, Rossman, M, Saenger, W. | | Deposit date: | 2007-09-18 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human saposins C and d: implications for lipid recognition and membrane interactions.
Structure, 16, 2008
|
|
3QP6
 
 | | Crystal structure of CviR (Chromobacterium violaceum 12472) bound to C6-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
2F84
 
 | |
5ND6
 
 | | Crystal structure of apo transketolase from Chlamydomonas reinhardtii | | Descriptor: | 1,2-ETHANEDIOL, Transketolase | | Authors: | Fermani, S, Zaffagnini, M, Francia, F, Pasquini, M. | | Deposit date: | 2017-03-07 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis for the magnesium-dependent activation of transketolase from Chlamydomonas reinhardtii.
Biochim. Biophys. Acta, 1861, 2017
|
|
3QP8
 
 | | Crystal structure of CviR (Chromobacterium violaceum 12472) ligand-binding domain bound to C10-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]decanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3QP5
 
 | | Crystal structure of CviR bound to antagonist chlorolactone (CL) | | Descriptor: | 4-(4-chlorophenoxy)-N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, CviR transcriptional regulator | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.249 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
2Q2A
 
 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ARGININE, ArtJ, SULFATE ION | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-26 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
3QP1
 
 | | Crystal structure of CviR ligand-binding domain bound to the native ligand C6-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
1G6W
 
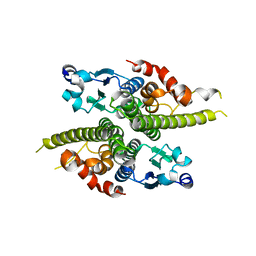 | | CRYSTAL STRUCTURE OF THE GLOBULAR REGION OF THE PRION PROTEIN URE2 FROM THE YEAST SACCAROMYCES CEREVISIAE | | Descriptor: | URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Janin, J, Melki, R, Morera, S. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the globular region of the prion protein Ure2 from the yeast Saccharomyces cerevisiae.
Structure, 9, 2001
|
|
2Q2C
 
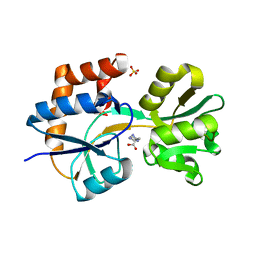 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ArtJ, GLYCEROL, HISTIDINE, ... | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-28 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
2PVU
 
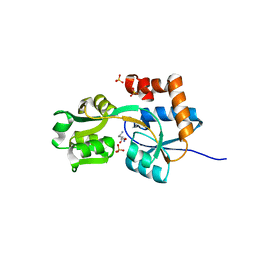 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ArtJ, LYSINE, SULFATE ION | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-10 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
1G6Y
 
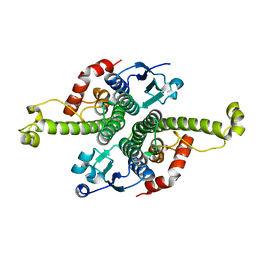 | | CRYSTAL STRUCTURE OF THE GLOBULAR REGION OF THE PRION PROTEIN URE2 FROM YEAST SACCHAROMYCES CEREVISIAE | | Descriptor: | URE2 PROTEIN | | Authors: | Bousset, L, Belrhali, H, Janin, J, Melki, R, Morera, S. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the globular region of the prion protein Ure2 from the yeast Saccharomyces cerevisiae.
Structure, 9, 2001
|
|
1O0N
 
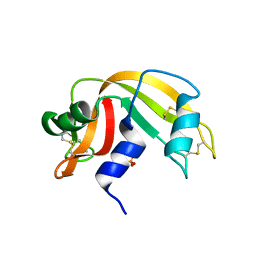 | | Ribonuclease A in complex with uridine-3'-phosphate | | Descriptor: | 3'-URIDINEMONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Leonidas, D.D, Oikonomakos, N.G, Chrysina, E.D, Kosmopoulou, M.N, Vlassi, M. | | Deposit date: | 2003-02-24 | | Release date: | 2003-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structures of ribonuclease A complexed with adenylic and uridylic nucleotide inhibitors. Implications for structure-based design of ribonucleolytic inhibitors
PROTEIN SCI., 12, 2003
|
|
1ZSO
 
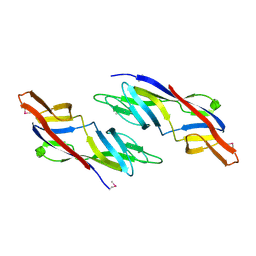 | |
3BO8
 
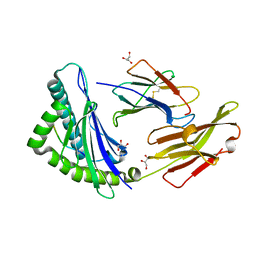 | | The High Resolution Crystal Structure of HLA-A1 Complexed with the MAGE-A1 Peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2007-12-17 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational changes within the HLA-A1:MAGE-A1 complex induced by binding of a recombinant antibody fragment with TCR-like specificity
Protein Sci., 18, 2009
|
|
1YZV
 
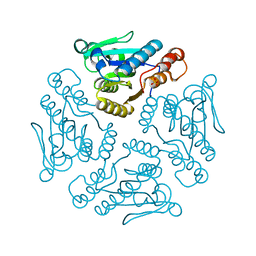 | |
4E44
 
 | |
1G27
 
 | | CRYSTAL STRUCTURE OF E.COLI POLYPEPTIDE DEFORMYLASE COMPLEXED WITH THE INHIBITOR BB-3497 | | Descriptor: | 2-[(FORMYL-HYDROXY-AMINO)-METHYL]-HEXANOIC ACID (1-DIMETHYLCARBAMOYL-2,2-DIMETHYL-PROPYL)-AMIDE, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-17 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
4E45
 
 | | Crystal structure of the hMHF1/hMHF2 Histone-Fold Tetramer in Complex with Fanconi Anemia Associated Helicase hFANCM | | Descriptor: | Centromere protein S, Centromere protein X, Fanconi anemia group M protein, ... | | Authors: | Fox III, D, Zhao, Y, Yang, W, Weidong, W. | | Deposit date: | 2012-03-12 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Reveal that FANCM remodels the MHF Tetramer in favor of binding Branched DNA
To be Published
|
|
1G2A
 
 | | THE CRYSTAL STRUCTURE OF E.COLI PEPTIDE DEFORMYLASE COMPLEXED WITH ACTINONIN | | Descriptor: | ACTINONIN, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-18 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
