4WMA
 
 | |
4WM0
 
 | |
4WN2
 
 | |
2RFC
 
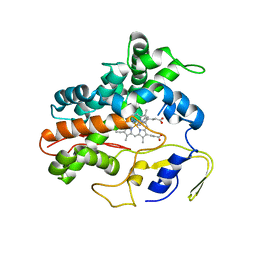 | | Ligand bound (4-phenylimidazole) Crystal Structure of a Cytochrome P450 from the Thermoacidophilic Archaeon Picrophilus Torridus | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ho, W.W, Li, H, Poulos, T.L, Nishida, C.R, Ortiz de Montellano, P.R. | | Deposit date: | 2007-09-28 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure and Properties of CYP231A2 from the Thermoacidophilic Archaeon Picrophilus torridus.
Biochemistry, 47, 2008
|
|
2RFB
 
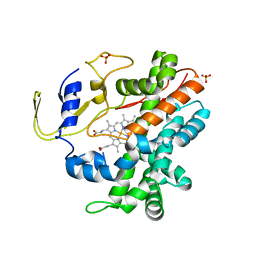 | | Crystal Structure of a Cytochrome P450 from the Thermoacidophilic Archaeon Picrophilus Torridus | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Ho, W.W, Li, H, Poulos, T.L, Nishida, C.R, Ortiz de Montellano, P.R. | | Deposit date: | 2007-09-28 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Properties of CYP231A2 from the Thermoacidophilic Archaeon Picrophilus torridus.
Biochemistry, 47, 2008
|
|
2RFK
 
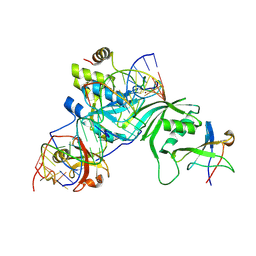 | | Substrate RNA Positioning in the Archaeal H/ACA Ribonucleoprotein Complex | | Descriptor: | Probable tRNA pseudouridine synthase B, Ribosome biogenesis protein Nop10, Small nucleolar rnp similar to gar1, ... | | Authors: | Liang, B, Xue, S, Terns, R.M, Terns, M.P, Li, H, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-09-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2RK5
 
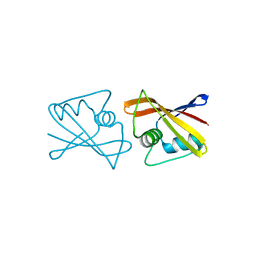 | |
1JFF
 
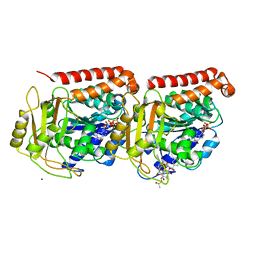 | | Refined structure of alpha-beta tubulin from zinc-induced sheets stabilized with taxol | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lowe, J, Li, H, Downing, K.H, Nogales, E. | | Deposit date: | 2001-06-20 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Refined structure of alpha beta-tubulin at 3.5 A resolution.
J.Mol.Biol., 313, 2001
|
|
1JCI
 
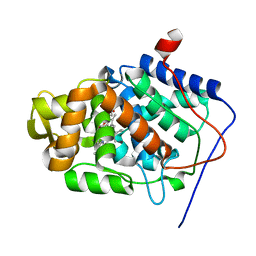 | | Stabilization of the Engineered Cation-binding Loop in Cytochrome c Peroxidase (CcP) | | Descriptor: | Cytochrome C Peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhaskar, B, Bonagura, C.A, Li, H, Poulos, T.L. | | Deposit date: | 2001-06-09 | | Release date: | 2002-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cation-induced stabilization of the engineered cation-binding loop in cytochrome c peroxidase (CcP).
Biochemistry, 41, 2002
|
|
1S4K
 
 | |
1DMK
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 4-AMINO-6-PHENYL-TETRAHYDROPTERIDINE | | Descriptor: | 2,4-DIAMINO-6-PHENYL-5,6,7,8,-TETRAHYDROPTERIDINE, ACETATE ION, CACODYLATE ION, ... | | Authors: | Kotsonis, P, Frohlich, L.G, Raman, C.S, Li, H, Berg, M, Gerwig, R, Groehn, V, Kang, Y, Al-Masoudi, N, Taghavi-Moghadam, S, Mohr, D, Munch, U, Schnabel, J, Martasek, P, Masters, B.S, Strobel, H, Poulos, T, Matter, H, Pfleiderer, W, Schmidt, H.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for pterin antagonism in nitric-oxide synthase. Development of novel 4-oxo-pteridine antagonists of (6R)-5,6,7,8-tetrahydrobiopterin
J.Biol.Chem., 276, 2001
|
|
4K5H
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with (S)-1,2-bis((2-amino-4-methylpyridin-6-yl)-methoxy)-propan-3-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-{[(2S)-3-aminopropane-1,2-diyl]bis(oxymethanediyl)}bis(4-methylpyridin-2-amine), ACETATE ION, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2013-04-14 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Chiral linkers to improve selectivity of double-headed neuronal nitric oxide synthase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1DMJ
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 5,6-CYCLIC-TETRAHYDROPTERIDINE | | Descriptor: | 7-AMINO-3,3A,4,5-TETRAHYDRO-8H-2-OXA-5,6,8,9B-TETRAAZA-CYCLOPENTA[A]NAPHTHALENE-1,9-DIONE, ACETATE ION, CACODYLATE ION, ... | | Authors: | Kotsonis, P, Frohlich, L.G, Raman, C.S, Li, H, Berg, M, Gerwig, R, Groehn, V, Kang, Y, Al-Masoudi, N, Taghavi-Moghadam, S, Mohr, D, Munch, U, Schnabel, J, Martasek, P, Masters, B.S, Strobel, H, Poulos, T, Matter, H, Pfleiderer, W, Schmidt, H.H. | | Deposit date: | 1999-12-14 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for pterin antagonism in nitric-oxide synthase. Development of novel 4-oxo-pteridine antagonists of (6R)-5,6,7,8-tetrahydrobiopterin
J.Biol.Chem., 276, 2001
|
|
6PTJ
 
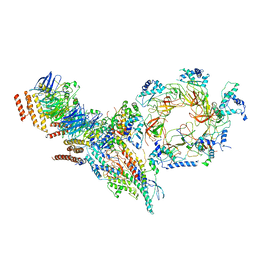 | | Structure of Ctf4 trimer in complex with one CMG helicase | | Descriptor: | Cell division control protein 45, DNA polymerase alpha-binding protein, DNA replication complex GINS protein PSF1, ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | Deposit date: | 2019-07-15 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PTO
 
 | | Structure of Ctf4 trimer in complex with three CMG helicases | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PSY
 
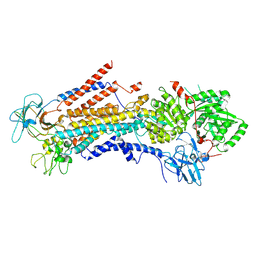 | | Cryo-EM structure of S. cerevisiae Drs2p-Cdc50p in the autoinhibited apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell division control protein 50, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2019-07-14 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p.
Nat Commun, 10, 2019
|
|
6PTN
 
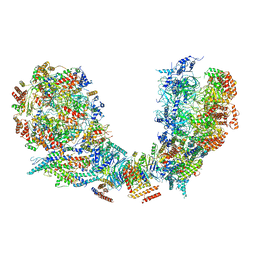 | | Structure of Ctf4 trimer in complex with two CMG helicases | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA polymerase alpha-binding protein, ... | | Authors: | Yuan, Z, Georgescu, R, Bai, L, Santos, R, Donnell, M, Li, H. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Ctf4 organizes sister replisomes and Pol alpha into a replication factory.
Elife, 8, 2019
|
|
6PSX
 
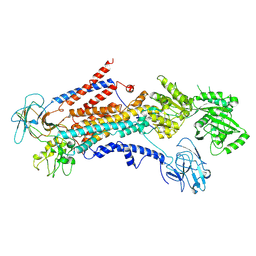 | | Cryo-EM structure of S. cerevisiae Drs2p-Cdc50p in the PI4P-activated form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cell division control protein 50, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2019-07-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Autoinhibition and activation mechanisms of the eukaryotic lipid flippase Drs2p-Cdc50p.
Nat Commun, 10, 2019
|
|
3LWR
 
 | | Structure of H/ACA RNP bound to a substrate RNA containing 4SU | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(4SU)P*GP*CP*GP*GP*UP*UP*U)-3', 50S ribosomal protein L7Ae, H/ACA RNA, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Functional and Structural Impact of Target Uridine Substitutions on the H/ACA Ribonucleoprotein Particle Pseudouridine Synthase .
Biochemistry, 49, 2010
|
|
3LWQ
 
 | | Structure of H/ACA RNP bound to a substrate RNA containing 3MU | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(UR3)P*GP*CP*GP*GP*UP*UP*U)-3, 50S ribosomal protein L7Ae, H/ACA RNA, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.678 Å) | | Cite: | Functional and Structural Impact of Target Uridine Substitutions on the H/ACA Ribonucleoprotein Particle Pseudouridine Synthase .
Biochemistry, 49, 2010
|
|
3M4R
 
 | | Structure of the N-terminal Class II Aldolase domain of a conserved protein from Thermoplasma acidophilum | | Descriptor: | CHLORIDE ION, Uncharacterized protein, ZINC ION | | Authors: | Cuff, M.E, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-11 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the N-terminal Class II Aldolase domain of a conserved protein from Thermoplasma acidophilum
TO BE PUBLISHED
|
|
3M91
 
 | |
3MR7
 
 | |
3MN2
 
 | |
3MQK
 
 | | Cbf5-Nop10-Gar1 complex binding with 17mer RNA containing ACA trinucleotide | | Descriptor: | RNA (5'-R(P*CP*GP*AP*UP*CP*CP*AP*CP*A)-3'), RNA (5'-R(P*GP*UP*UP*CP*GP*AP*UP*CP*CP*AP*CP*AP*G)-3'), Ribosome biogenesis protein Nop10, ... | | Authors: | Zhou, J, Liang, B, Li, H. | | Deposit date: | 2010-04-28 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional evidence of high specificity of Cbf5 for ACA trinucleotide.
Rna, 17, 2011
|
|
