6LE2
 
 | | Structure of D-carbamoylase mutant from Nitratireductor indicus | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-carbamoyl-D-amino-acid hydrolase | | Authors: | Ni, Y, Liu, Y.F, Xu, G.C, Dai, W. | | Deposit date: | 2019-11-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure-Guided Engineering of D-Carbamoylase Reveals a Key Loop at Substrate Entrance Tunnel
Acs Catalysis, 10, 2020
|
|
5OID
 
 | | Complex Trichoplax STIL-NTD:human CEP85 coiled coil domain 4 | | Descriptor: | Centrosomal protein of 85 kDa, Putative uncharacterized protein | | Authors: | van Breugel, M. | | Deposit date: | 2017-07-18 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Direct binding of CEP85 to STIL ensures robust PLK4 activation and efficient centriole assembly.
Nat Commun, 9, 2018
|
|
5OI9
 
 | | Trichoplax adhaerens STIL N-terminal domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative uncharacterized protein | | Authors: | van Breugel, M. | | Deposit date: | 2017-07-18 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Direct binding of CEP85 to STIL ensures robust PLK4 activation and efficient centriole assembly.
Nat Commun, 9, 2018
|
|
5OI7
 
 | | Human CEP85 - coiled coil domain 4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Centrosomal protein of 85 kDa | | Authors: | van Breugel, M. | | Deposit date: | 2017-07-18 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Direct binding of CEP85 to STIL ensures robust PLK4 activation and efficient centriole assembly.
Nat Commun, 9, 2018
|
|
6LHG
 
 | | Crystal structure of chicken cCD8aa/pBF2*04:01 | | Descriptor: | Beta-2-microglobulin, CD8 alpha chain, IE8 peptide, ... | | Authors: | Liu, Y.J, Xia, C. | | Deposit date: | 2019-12-08 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Combination of CD8 alpha alpha and Peptide-MHC-I in a Face-to-Face Mode Promotes Chicken gamma delta T Cells Response.
Front Immunol, 11, 2020
|
|
7VJQ
 
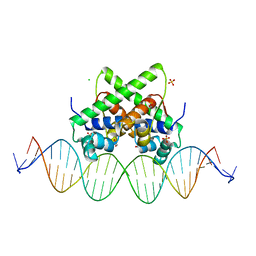 | | Pectobacterium phage ZF40 apo-aca2 complexed with 26bp DNA substrate | | Descriptor: | CHLORIDE ION, DNA (27-MER), GLYCEROL, ... | | Authors: | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7VJO
 
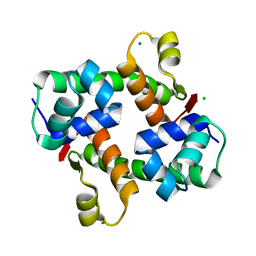 | | Pectobacterium phage ZF40 apo-Aca2 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, anti-CRISPR-associated protein Aca2 | | Authors: | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7VJN
 
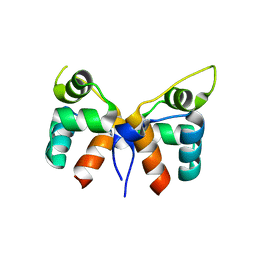 | |
7VJM
 
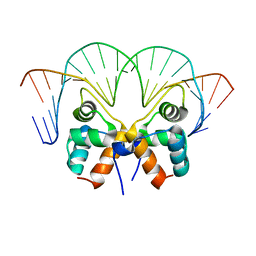 | | Aca1 in complex with 19bp palindromic DNA substrate | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*CP*AP*CP*AP*TP*TP*GP*TP*GP*CP*CP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*GP*GP*CP*AP*CP*AP*AP*TP*GP*TP*GP*CP*CP*TP*AP*A)-3'), anti-CRISPR-associated protein Aca1 | | Authors: | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7VJP
 
 | | Selenomethionine-derived Pectobacterium phage ZF40 apo-Aca2 | | Descriptor: | SULFATE ION, anti-CRISPR-associated protein Aca2 | | Authors: | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | Deposit date: | 2021-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7KDC
 
 | | The complex between RhoD and the Plexin B2 RBD | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-B2, ... | | Authors: | Kuo, Y, Wang, Y, Zhang, x. | | Deposit date: | 2020-10-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A putative structural mechanism underlying the antithetic effect of homologous RND1 and RhoD GTPases in mammalian plexin regulation.
Elife, 10, 2021
|
|
6V9T
 
 | | Tudor domain of TDRD3 in complex with a small molecule | | Descriptor: | 4-methyl-2,3,4,5,6,7-hexahydrodicyclopenta[b,e]pyridin-8(1H)-imine, Tudor domain-containing protein 3, UNKNOWN ATOM OR ION | | Authors: | Li, W, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-16 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | A small molecule antagonist of SMN disrupts the interaction between SMN and RNAP II.
Nat Commun, 13, 2022
|
|
5UY1
 
 | | X-ray crystal structure of apo Halotag | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Dunham, N.P, Boal, A.K. | | Deposit date: | 2017-02-23 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Cation-pi Interaction Enables a Halo-Tag Fluorogenic Probe for Fast No-Wash Live Cell Imaging and Gel-Free Protein Quantification.
Biochemistry, 56, 2017
|
|
5UXZ
 
 | |
5CNQ
 
 | | Crystal structure of the Holliday junction-resolving enzyme GEN1 (WT) in complex with product DNA, Mg2+ and Mn2+ ions | | Descriptor: | DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MANGANESE (II) ION, Nuclease-like protein, ... | | Authors: | Liu, Y.J, Freeman, A.D.J, Declais, A.C, Wilson, T.J, Gartner, A, Lilley, D.M.J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Cell Rep, 13, 2015
|
|
5CO8
 
 | | Crystal structure of the Holliday junction-resolving enzyme GEN1 (WT) in complex with product DNA and Mg2+ ion | | Descriptor: | DNA (31-MER), DNA (5'-D(*AP*GP*AP*CP*TP*GP*CP*AP*GP*TP*TP*GP*AP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*A)-3'), ... | | Authors: | Liu, Y.J, Freeman, A.D.J, Declais, A.C, Wilson, T.J, Gartner, A, Lilley, D.M.J. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a Eukaryotic GEN1 Resolving Enzyme Bound to DNA.
Cell Rep, 13, 2015
|
|
5EB9
 
 | | Crystal Structure Of Chicken CD8aa Homodimer | | Descriptor: | CD8 alpha chain | | Authors: | Liu, Y.J, Qi, J.X, Xia, C. | | Deposit date: | 2015-10-18 | | Release date: | 2016-09-14 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | The structural basis of chicken, swine and bovine CD8 alpha alpha dimers provides insight into the co-evolution with MHC I in endotherm species.
Sci Rep, 6, 2016
|
|
6XZ9
 
 | | Structure of aldosterone synthase (CYP11B2) in complex with 5-chloro-3,3-dimethyl-2-[5-[1-(1-methylpyrazole-4-carbonyl)azetidin-3-yl]oxy-3-pyridyl]isoindolin-1-one | | Descriptor: | 5-chloranyl-3,3-dimethyl-2-[5-[1-(1-methylpyrazol-4-yl)carbonylazetidin-3-yl]oxypyridin-3-yl]isoindol-1-one, Cytochrome P450 11B2, mitochondrial, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
6XZ8
 
 | | Structure of aldosterone synthase (CYP11B2) in complex with N-[(1R)-1-[5-(6-chloro-1,1-dimethyl-3-oxo-isoindolin-2-yl)-3-pyridyl]ethyl]methanesulfonamide | | Descriptor: | Cytochrome P450 11B2, mitochondrial, HEME C, ... | | Authors: | Kuglstatter, A, Joseph, C, Benz, J. | | Deposit date: | 2020-02-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of 3-Pyridyl Isoindolin-1-one Derivatives as Potent, Selective, and Orally Active Aldosterone Synthase (CYP11B2) Inhibitors.
J.Med.Chem., 63, 2020
|
|
5EDX
 
 | | Crystal structure of swine CD8aa homodimer | | Descriptor: | CD8 alpha antigen | | Authors: | Liu, Y.J, Qi, J.X, Zhang, N.Z, Xia, C. | | Deposit date: | 2015-10-22 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The structural basis of chicken, swine and bovine CD8 alpha alpha dimers provides insight into the co-evolution with MHC I in endotherm species.
Sci Rep, 6, 2016
|
|
1KVU
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
7RKA
 
 | |
1HSJ
 
 | | SARR MBP FUSION STRUCTURE | | Descriptor: | FUSION PROTEIN CONSISTING OF STAPHYLOCOCCUS ACCESSORY REGULATOR PROTEIN R AND MALTOSE BINDING PROTEIN, alpha-D-glucopyranose | | Authors: | Zhang, G. | | Deposit date: | 2000-12-26 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the SarR protein from Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
4FGX
 
 | | Crystal structure of bace1 with novel inhibitor | | Descriptor: | Beta-secretase 1, DI(HYDROXYETHYL)ETHER, Inhibitor (2R,5S,8S,12S,13S,16S,19S,22S)-16-(3-amino-3-oxopropyl)-2,13-dibenzyl-12,22-dihydroxy-8-isobutyl-19-isopropyl-3,5,17-trimethyl-4,7,10,15,18,21-hexaoxo-3,6,9,14,17,20-hexaazatricosan-1-oic acid, ... | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2012-06-05 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
6AGS
 
 | |
