2CWH
 
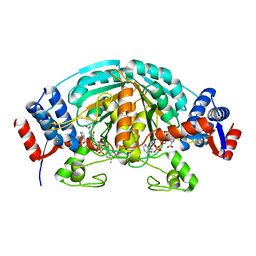 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae complexed with NADPH and pyrrole-2-carboxylate | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRROLE-2-CARBOXYLATE, delta1-piperideine-2-carboxylate reductase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2005-06-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
2CWF
 
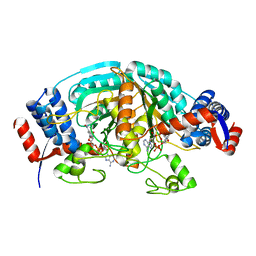 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, delta1-piperideine-2-carboxylate reductase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2005-06-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
2DQ4
 
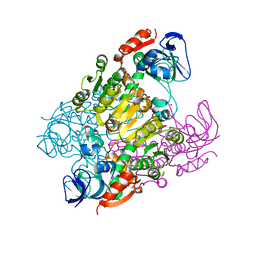 | | Crystal structure of threonine 3-dehydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, L-threonine 3-dehydrogenase, ZINC ION | | Authors: | Omi, R, Yao, T, Goto, M, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-19 | | Release date: | 2007-06-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of threonine 3-dehydrogenase
To be Published
|
|
1ET0
 
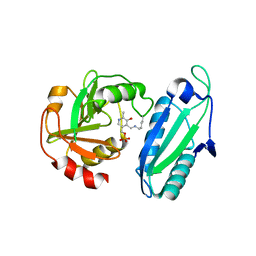 | | CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Nakai, T, Mizutani, H, Miyahara, I, Hirotsu, K, Takeda, S, Jhee, K.H, Yoshimura, T, Esaki, N. | | Deposit date: | 2000-04-12 | | Release date: | 2000-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of 4-amino-4-deoxychorismate lyase from Escherichia coli.
J.Biochem.(Tokyo), 128, 2000
|
|
1G7X
 
 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R292L/R386L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1G4V
 
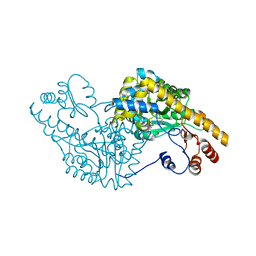 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/Y225F | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-10-28 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1G7W
 
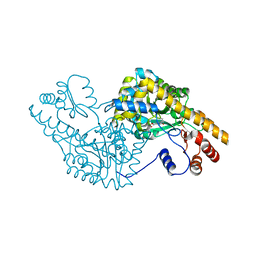 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R386L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1GEX
 
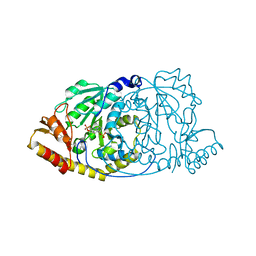 | | CRYSTAL STRUCTURE OF HISTIDINOL-PHOSPHATE AMINOTRANSFERASE COMPLEXED WITH HISTIDINOL-PHOSPHATE | | Descriptor: | HISTIDINOL-PHOSPHATE AMINOTRANSFERASE, PHOSPHORIC ACID MONO-[2-AMINO-3-(3H-IMIDAZOL-4-YL)-PROPYL]ESTER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Haruyama, K, Nakai, T, Miyahara, I, Hirotsu, K, Mizuguchi, H, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5'-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
1GEY
 
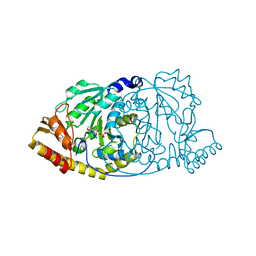 | | CRYSTAL STRUCTURE OF HISTIDINOL-PHOSPHATE AMINOTRANSFERASE COMPLEXED WITH N-(5'-PHOSPHOPYRIDOXYL)-L-GLUTAMATE | | Descriptor: | 4-[(1,3-DICARBOXY-PROPYLAMINO)-METHYL]-3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDINIUM, HISTIDINOL-PHOSPHATE AMINOTRANSFERASE | | Authors: | Haruyama, K, Nakai, T, Miyahara, I, Hirotsu, K, Mizuguchi, H, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5'-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
1GEW
 
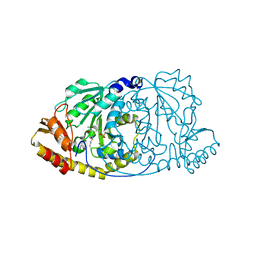 | | CRYSTAL STRUCTURE OF HISTIDINOL-PHOSPHATE AMINOTRANSFERASE COMPLEXED WITH PYRIDOXAL 5'-PHOSPHATE | | Descriptor: | HISTIDINOL-PHOSPHATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Haruyama, K, Nakai, T, Miyahara, I, Hirotsu, K, Mizuguchi, H, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5'-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
1EVI
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE PURPLE INTERMEDIATE OF PORCINE KIDNEY D-AMINO ACID OXIDASE | | Descriptor: | 3,4-DIHYDRO-2H-PYRROLIUM-5-CARBOXYLATE, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Mizutani, H, Miyahara, I, Hirotsu, K, Nishina, Y, Shiga, K, Setoyama, C, Miura, R. | | Deposit date: | 2000-04-20 | | Release date: | 2000-10-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of the purple intermediate of porcine kidney D-amino acid oxidase. Optimization of the oxidative half-reaction through alignment of the product with reduced flavin.
J.Biochem.(Tokyo), 128, 2000
|
|
1UDY
 
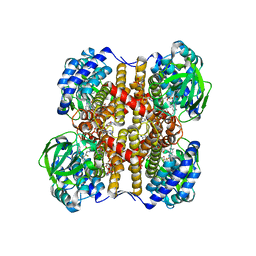 | | Medium-Chain Acyl-CoA Dehydrogenase with 3-Thiaoctanoyl-CoA | | Descriptor: | 3-THIAOCTANOYL-COENZYME A, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Satoh, A, Nakajima, Y, Miyahara, I, Hirotsu, K, Tanaka, T, Nishina, Y, Shiga, K, Tamaoki, H, Setoyama, C, Miura, R. | | Deposit date: | 2003-05-07 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the transition state analog of medium-chain acyl-CoA dehydrogenase. Crystallographic and molecular orbital studies on the charge-transfer complex of medium-chain acyl-CoA dehydrogenase with 3-thiaoctanoyl-CoA
J.BIOCHEM.(TOKYO), 134, 2003
|
|
1VE9
 
 | | Porcine kidney D-amino acid oxidase | | Descriptor: | BENZOIC ACID, D-amino acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Mizutani, H, Miyahara, I, Hirotsu, K, Nishina, Y, Shiga, K, Setoyama, C, Miura, R. | | Deposit date: | 2004-03-29 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of porcine kidney D-amino acid oxidase at 3.0 A resolution.
J.Biochem., 120, 1996
|
|
1VEF
 
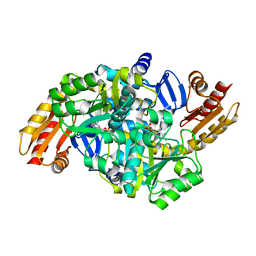 | | Acetylornithine aminotransferase from Thermus thermophilus HB8 | | Descriptor: | Acetylornithine/acetyl-lysine aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-30 | | Release date: | 2005-08-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Three-Dimensional Strutcure of Acetylornithine aminotransferase from Thermus thermophilus HB8
To be Published
|
|
1V93
 
 | | 5,10-Methylenetetrahydrofolate Reductase from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5,10-Methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Nakajima, Y, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-20 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of 5,10-Methylenetetrahydrofolate reductase from Thermus thermophilus HB8
To be Published
|
|
1WTJ
 
 | | Crystal Structure of delta1-piperideine-2-carboxylate reductase from Pseudomonas syringae pvar.tomato | | Descriptor: | ureidoglycolate dehydrogenase | | Authors: | Goto, M, Muramatsu, H, Mihara, H, Kurihara, T, Esaki, N, Omi, R, Miyahara, I, Hirotsu, K. | | Deposit date: | 2004-11-24 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of Delta1-piperideine-2-carboxylate/Delta1-pyrroline-2-carboxylate reductase belonging to a new family of NAD(P)H-dependent oxidoreductases: conformational change, substrate recognition, and stereochemistry of the reaction
J.Biol.Chem., 280, 2005
|
|
1WTC
 
 | | Crystal Structure of S.pombe Serine Racemase complex with AMPPCP | | Descriptor: | Hypothetical protein C320.14 in chromosome III, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Goto, M, Miyahara, I, Hirotsu, K. | | Deposit date: | 2004-11-22 | | Release date: | 2005-11-01 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a homolog of mammalian serine racemase from Schizosaccharomyces pombe
J.Biol.Chem., 284, 2009
|
|
1WKH
 
 | | Acetylornithine aminotransferase from thermus thermophilus HB8 | | Descriptor: | 4-[(1,3-DICARBOXY-PROPYLAMINO)-METHYL]-3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDINIUM, Acetylornithine/acetyl-lysine aminotransferase | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2005-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Acetylornithine aminotransferase from thermus thermophilus HB8
To be Published
|
|
1WKG
 
 | | Acetylornithine aminotransferase from thermus thermophilus HB8 | | Descriptor: | Acetylornithine/acetyl-lysine aminotransferase, N~2~-ACETYL-N~5~-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-ORNITHINE | | Authors: | Matsumura, M, Goto, M, Omi, R, Miyahara, I, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-31 | | Release date: | 2005-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Acetylornithine aminotransferase from thermus thermophilus HB8
To be Published
|
|
1BKG
 
 | | ASPARTATE AMINOTRANSFERASE FROM THERMUS THERMOPHILUS WITH MALEATE | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, MALEIC ACID | | Authors: | Nakai, T, Okada, K, Kuramitsu, S, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-07-07 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Thermus thermophilus HB8 aspartate aminotransferase and its complex with maleate.
Biochemistry, 38, 1999
|
|
1BJW
 
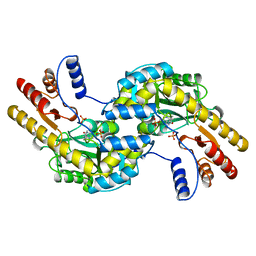 | | ASPARTATE AMINOTRANSFERASE FROM THERMUS THERMOPHILUS | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PHOSPHATE ION | | Authors: | Nakai, T, Okada, K, Kuramitsu, S, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-06-30 | | Release date: | 1999-07-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Thermus thermophilus HB8 aspartate aminotransferase and its complex with maleate.
Biochemistry, 38, 1999
|
|
1GC3
 
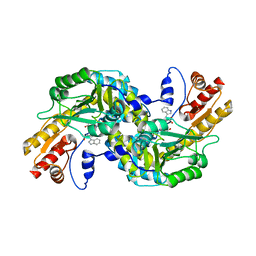 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE TETRA MUTANT 2 COMPLEXED WITH TRYPTOPHAN | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN | | Authors: | Ura, H, Nakai, T, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 2000-07-18 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme.
J.Biochem., 130, 2001
|
|
1GCK
 
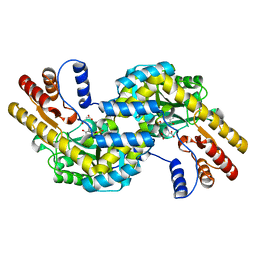 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE DOUBLE MUTANT 1 COMPLEXED WITH ASPARTATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, ASPARTIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 2000-08-04 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme.
J.Biochem., 130, 2001
|
|
1GC4
 
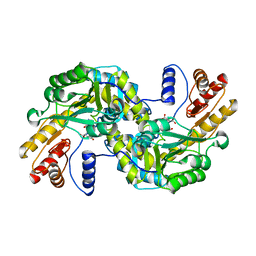 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE TETRA MUTANT 2 COMPLEXED WITH ASPARTATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, ASPARTIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 2000-07-19 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme.
J.Biochem., 130, 2001
|
|
1J1Z
 
 | |
