1TOL
 
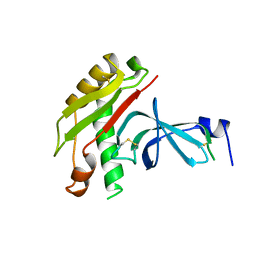 | | FUSION OF N-TERMINAL DOMAIN OF THE MINOR COAT PROTEIN FROM GENE III IN PHAGE M13, AND C-TERMINAL DOMAIN OF E. COLI PROTEIN-TOLA | | Descriptor: | PROTEIN (FUSION PROTEIN CONSISTING OF MINOR COAT PROTEIN, GLYCINE RICH LINKER, TOLA, ... | | Authors: | Lubkowski, J, Wlodawer, A, Hennecke, F, Plueckthun, A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Filamentous phage infection: crystal structure of g3p in complex with its coreceptor, the C-terminal domain of TolA.
Structure Fold.Des., 7, 1999
|
|
5L04
 
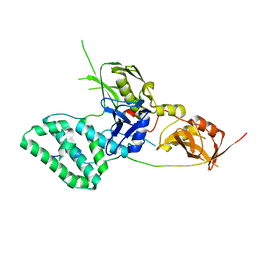 | |
5FIV
 
 | | STRUCTURAL STUDIES OF HIV AND FIV PROTEASES COMPLEXED WITH AN EFFICIENT INHIBITOR OF FIV PR | | Descriptor: | RETROPEPSIN, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Li, M, Lee, T, Morris, G, Laco, G, Wong, C, Olson, A, Elder, J, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-12-02 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease
Proteins, 38, 2000
|
|
1AGX
 
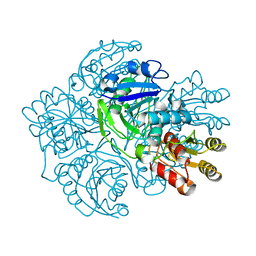 | | REFINED CRYSTAL STRUCTURE OF ACINETOBACTER GLUTAMINASIFICANS GLUTAMINASE-ASPARAGINASE | | Descriptor: | GLUTAMINASE-ASPARAGINASE | | Authors: | Lubkowski, J, Wlodawer, A, Housset, D, Weber, I.T, Ammon, H.L, Murphy, K.C, Swain, A.L. | | Deposit date: | 1994-07-13 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refined crystal structure of Acinetobacter glutaminasificans glutaminase-asparaginase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
8DVH
 
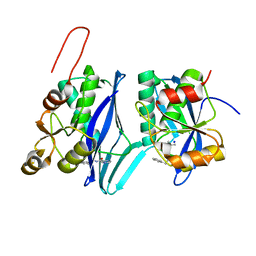 | | Crystal structure of ATP-dependent Lon protease from Bacillus subtillis (BsLonBA) | | Descriptor: | Lon protease 2, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, SODIUM ION | | Authors: | Sekula, B, Li, M, Gustchina, A, Wlodawer, A. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unique Structural Fold of LonBA Protease from Bacillus subtilis, a Member of a Newly Identified Subfamily of Lon Proteases.
Int J Mol Sci, 23, 2022
|
|
7HVP
 
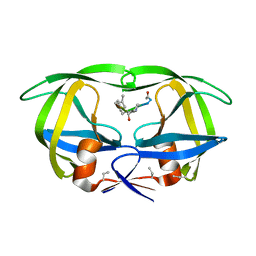 | | X-RAY CRYSTALLOGRAPHIC STRUCTURE OF A COMPLEX BETWEEN A SYNTHETIC PROTEASE OF HUMAN IMMUNODEFICIENCY VIRUS 1 AND A SUBSTRATE-BASED HYDROXYETHYLAMINE INHIBITOR | | Descriptor: | HIV-1 PROTEASE, INHIBITOR ACE-SER-LEU-ASN-PHE-PSI(CH(OH)-CH2N)-PRO-ILE VME (JG-365) | | Authors: | Swain, A.L, Miller, M.M, Green, J, Rich, D.H, Schneider, J, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1990-09-13 | | Release date: | 1993-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic structure of a complex between a synthetic protease of human immunodeficiency virus 1 and a substrate-based hydroxyethylamine inhibitor.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
3PGA
 
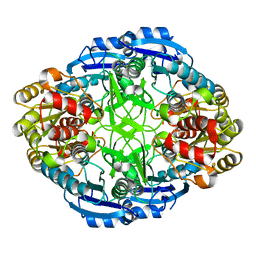 | | STRUCTURAL CHARACTERIZATION OF PSEUDOMONAS 7A GLUTAMINASE-ASPARAGINASE | | Descriptor: | GLUTAMINASE-ASPARAGINASE | | Authors: | Lubkowski, J, Wlodawer, A, Ammon, H.L, Copeland, T.D, Swain, A.L. | | Deposit date: | 1994-07-19 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of Pseudomonas 7A glutaminase-asparaginase.
Biochemistry, 33, 1994
|
|
7UPZ
 
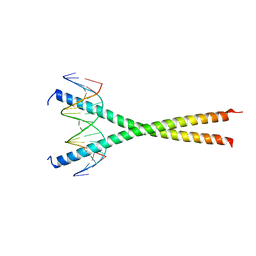 | | Structural basis for cell type specific DNA binding of C/EBPbeta: the case of cell cycle inhibitor p15INK4b promoter | | Descriptor: | CCAAT/enhancer-binding protein beta, DNA (5'-D(*AP*TP*TP*CP*TP*TP*AP*AP*GP*AP*AP*AP*GP*AP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*TP*CP*TP*TP*TP*CP*TP*TP*AP*AP*GP*AP*A)-3') | | Authors: | Lountos, G.T, Cherry, S, Tropea, J.E, Wlodawer, A, Miller, M. | | Deposit date: | 2022-04-18 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural basis for cell type specific DNA binding of C/EBP beta : The case of cell cycle inhibitor p15INK4b promoter.
J.Struct.Biol., 214, 2022
|
|
7UT3
 
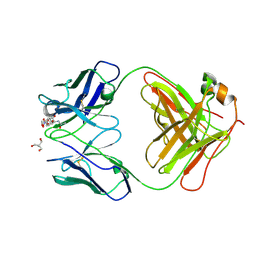 | | Crystal structure of complex of Fab, G10C with GalNAc-pNP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-nitrophenyl 2-acetamido-2-deoxy-alpha-D-galactopyranoside, Fab protein heavy chain, ... | | Authors: | Li, M, Wlodawer, A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Development of a GalNAc-Tyrosine-Specific Monoclonal Antibody and Detection of Tyrosine O -GalNAcylation in Numerous Human Tissues and Cell Lines.
J.Am.Chem.Soc., 144, 2022
|
|
1NAG
 
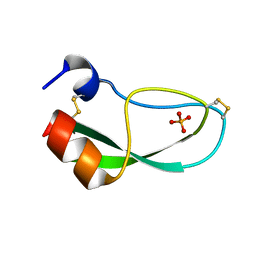 | | CREVICE-FORMING MUTANTS IN THE RIGID CORE OF BOVINE PANCREATIC TRYPSIN INHIBITOR: CRYSTAL STRUCTURES OF F22A, Y23A, N43G, AND F45A | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, PHOSPHATE ION | | Authors: | Danishefsky, A.T, Wlodawer, A, Kim, K.-S, Tao, F, Woodward, C. | | Deposit date: | 1992-08-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crevice-forming mutants in the rigid core of bovine pancreatic trypsin inhibitor: crystal structures of F22A, Y23A, N43G, and F45A.
Protein Sci., 2, 1993
|
|
4HVP
 
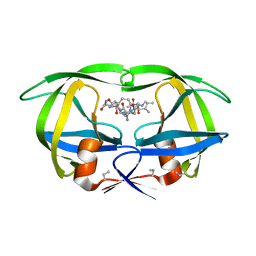 | | Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 Angstroms resolution | | Descriptor: | HIV-1 PROTEASE, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide | | Authors: | Miller, M, Schneider, J, Sathyanarayana, B.K, Toth, M.V, Marshall, G.R, Clawson, L, Selk, L, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1989-08-08 | | Release date: | 1990-04-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution.
Science, 246, 1989
|
|
9HFT
 
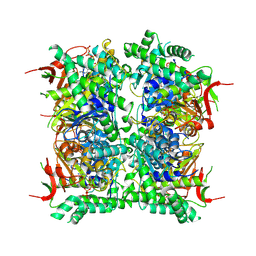 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): hexamer with 5mdCTP bound in the active site | | Descriptor: | Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION, [[(2~{R},3~{S},5~{R})-5-(4-azanyl-5-methyl-2-oxidanylidene-pyrimidin-1-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activity and structure of human (d)CTP deaminase CDADC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9HFQ
 
 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): trimer without a ligand | | Descriptor: | Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Activity and structure of human (d)CTP deaminase CDADC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9HFS
 
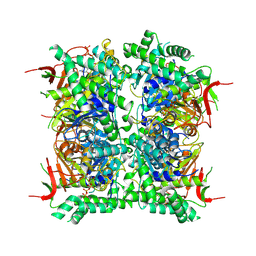 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): hexamer with dCTP bound in the active site | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Activity and structure of human (d)CTP deaminase CDADC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9HFR
 
 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): hexamer without a ligand | | Descriptor: | Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Activity and structure of human (d)CTP deaminase CDADC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9NWB
 
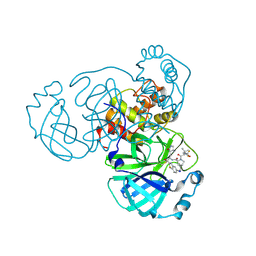 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-280-5I | | Descriptor: | (1R,2S,5S)-N-{(1S,2R)-1-hydroxy-1-(5-iodo-1,3-benzothiazol-2-yl)-3-[(3R)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, ORF1a polyprotein | | Authors: | Bulut, H, Hayashi, H, Hattori, S, Li, M, Das, D, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2025-03-21 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Impact of Single Halogen Atom Substitutions on Potency of Inhibitors Targeting SARS-CoV-2 Main Protease
To Be Published
|
|
9NWA
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-277-5Cl | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-chloro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Kuwata, N, Hayashi, H, Aoki, H, Das, D, Li, M, Wlodawer, A, Misumi, S, Tamamura, H, Mitsuya, H. | | Deposit date: | 2025-03-21 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Impact of Single Halogen Atom Substitutions on Potency of Inhibitors Targeting SARS-CoV-2 Main Protease
To Be Published
|
|
9NWC
 
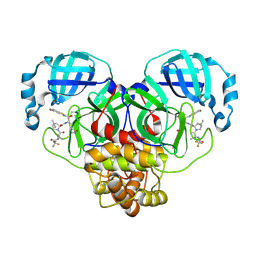 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-276-5Br | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-bromo-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-D-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Kuwata, N, Hayashi, H, Aoki, H, Hattori, S, Li, M, Das, D, Misumi, S, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2025-03-21 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Impact of Single Halogen Atom Substitutions on Potency of Inhibitors Targeting SARS-CoV-2 Main Protease
To Be Published
|
|
2HYQ
 
 | |
2HYR
 
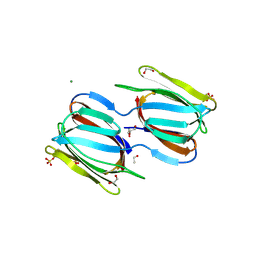 | | Crystal structure of a complex of griffithsin with maltose | | Descriptor: | 1,2-ETHANEDIOL, Griffithsin, MAGNESIUM ION, ... | | Authors: | Ziolkowska, N.E, Wlodawer, A. | | Deposit date: | 2006-08-07 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystallographic, thermodynamic, and molecular modeling studies of the mode of binding of oligosaccharides to the potent antiviral protein griffithsin.
Proteins, 67, 2007
|
|
4MON
 
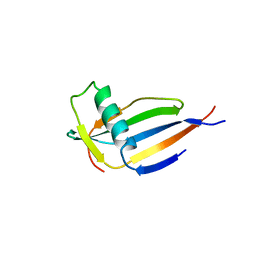 | | ORTHORHOMBIC MONELLIN | | Descriptor: | MONELLIN | | Authors: | Bujacz, G, Wlodawer, A. | | Deposit date: | 1997-03-04 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of monellin refined to 2.3 a resolution in the orthorhombic crystal form.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
9IX8
 
 | | Crystallization and structural characterization of phosphopentomutase from the hyperthermophilic archaeon Thermococcus kodakarensis | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, GLYCEROL, Phosphopentomutase | | Authors: | Naz, Z, Lubkowski, T.J, Saleem, M, Rahman, M, Wlodawer, A, Rashid, N. | | Deposit date: | 2024-07-26 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Biophysical Characterization of a Novel Phosphopentomutase from the Hyperthermophilic Archaeon Thermococcus kodakarensis .
Int J Mol Sci, 25, 2024
|
|
3FIV
 
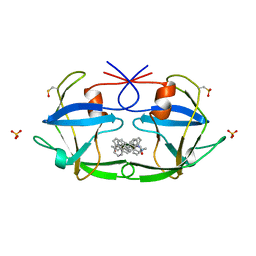 | | CRYSTAL STRUCTURE OF FELINE IMMUNODEFICIENCY VIRUS PROTEASE COMPLEXED WITH A SUBSTRATE | | Descriptor: | ACE-ALN-VAL-LEU-ALA-GLU-ALN-NH2, FELINE IMMUNODEFICIENCY VIRUS PROTEASE, SULFATE ION | | Authors: | Schalk-Hihi, C, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1997-07-09 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the inactive D30N mutant of feline immunodeficiency virus protease complexed with a substrate and an inhibitor.
Biochemistry, 36, 1997
|
|
4O1S
 
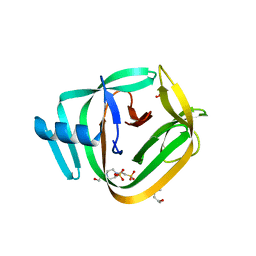 | | Crystal structure of TvoVMA intein | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, ... | | Authors: | Aranko, A.S, Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7008 Å) | | Cite: | Structure-based engineering and comparison of novel split inteins for protein ligation.
Mol Biosyst, 10, 2014
|
|
4O1R
 
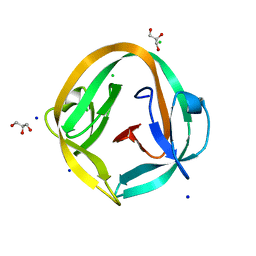 | | Crystal structure of NpuDnaB intein | | Descriptor: | CHLORIDE ION, GLYCEROL, Replicative DNA helicase, ... | | Authors: | Aranko, A.S, Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based engineering and comparison of novel split inteins for protein ligation.
Mol Biosyst, 10, 2014
|
|
