2X3A
 
 | | AsaP1 inactive mutant E294Q, an extracellular toxic zinc metalloendopeptidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Bogdanovic, X, Palm, G.J, Singh, R.K, Hinrichs, W. | | Deposit date: | 2010-01-22 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Evidence of Intramolecular Propeptide Inhibition of the Aspzincin Metalloendopeptidase Asap1.
FEBS Lett., 590, 2016
|
|
2X3B
 
 | | AsaP1 inactive mutant E294A, an extracellular toxic zinc metalloendopeptidase | | Descriptor: | TOXIC EXTRACELLULAR ENDOPEPTIDASE, ZINC ION | | Authors: | Bogdanovic, X, Palm, G.J, Singh, R.K, Hinrichs, W. | | Deposit date: | 2010-01-22 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Evidence of Intramolecular Propeptide Inhibition of the Aspzincin Metalloendopeptidase Asap1.
FEBS Lett., 590, 2016
|
|
2X3C
 
 | | AsaP1 inactive mutant E294Q, an extracellular toxic zinc metalloendopeptidase | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Bogdanovic, X, Palm, G.J, Singh, R.K, Hinrichs, W. | | Deposit date: | 2010-01-22 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Evidence of Intramolecular Propeptide Inhibition of the Aspzincin Metalloendopeptidase Asap1.
FEBS Lett., 590, 2016
|
|
2H8H
 
 | | Src kinase in complex with a quinazoline inhibitor | | Descriptor: | N-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-7-[2-(4-METHYLPIPERAZIN-1-YL)ETHOXY]-5-(TETRAHYDRO-2H-PYRAN-4-YLOXY)QUINAZOLIN-4-AMINE, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Otterbein, L.R, Norman, R, Pauptit, R.A, Rowsell, S, Breed, J. | | Deposit date: | 2006-06-07 | | Release date: | 2006-11-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | N-(5-chloro-1,3-benzodioxol-4-yl)-7-[2-(4-methylpiperazin-1-yl)ethoxy]-5- (tetrahydro-2H-pyran-4-yloxy)quinazolin-4-amine, a novel, highly selective, orally available, dual-specific c-Src/Abl kinase inhibitor.
J.Med.Chem., 49, 2006
|
|
3U4E
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain CAP45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG9 Heavy Chain, PG9 Light Chain, ... | | Authors: | Gorman, J, McLellan, J, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3TCL
 
 | | Crystal Structure of HIV-1 Neutralizing Antibody CH04 | | Descriptor: | CH04 Heavy Chain Fab, CH04 Light Chain Fab, IMIDAZOLE | | Authors: | Louder, R.K, McLellan, J.S, Pancera, M, Yang, Y, Zhang, B, Bonsignori, M, Kwong, P.D. | | Deposit date: | 2011-08-09 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U1S
 
 | | Crystal structure of human Fab PGT145, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | Fab PGT145 Heavy chain, Fab PGT145 Light chain, GLYCEROL, ... | | Authors: | Julien, J.-P, Diwanji, D, Burton, D.R, Wilson, I.A. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
2WQB
 
 | | Structure of the Tie2 kinase domain in complex with a thiazolopyrimidine inhibitor | | Descriptor: | 2-[3-(CYCLOHEXYLMETHYL)-5-PHENYL-IMIDAZOL-4-YL]-[1,3]THIAZOLO[4,5-E]PYRIMIDIN-7-AMINE, ANGIOPOIETIN-1 RECEPTOR | | Authors: | Brassington, C, Breed, J, Buttar, D, Fitzek, M, Forder, C, Hassall, L, Hayter, B.R, Jones, C.D, Luke, R.W.A, McCall, E, McCoull, W, Norman, R, Paterson, D, McMiken, H, Rowsell, S, Tucker, J.A. | | Deposit date: | 2009-08-18 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Novel Thienopyrimidine and Thiazolopyrimidine Kinase Inhibitors with Activity Against Tie-2 in Vitro and in Vivo.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4IUD
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form with ascorbate - partly reduced state | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Hammer, M, Schmidt, A, Frielingsdorf, S, Fritsch, J, Lenz, O, Scheerer, P. | | Deposit date: | 2013-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase.
Nat.Chem.Biol., 10, 2014
|
|
4IUC
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form - oxidized state 2 | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Frielingsdorf, S, Schmidt, A, Fritsch, J, Lenz, O, Scheerer, P. | | Deposit date: | 2013-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase.
Nat.Chem.Biol., 10, 2014
|
|
4IUB
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form - oxidized state 1 | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Frielingsdorf, S, Schmidt, A, Fritsch, J, Lenz, O, Scheerer, P. | | Deposit date: | 2013-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase.
Nat.Chem.Biol., 10, 2014
|
|
3U36
 
 | | Crystal Structure of PG9 Fab | | Descriptor: | PG9 Fab heavy chain, PG9 Fab light chain, SULFATE ION | | Authors: | McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.281 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U4B
 
 | | CH04H/CH02L Fab P4 | | Descriptor: | CH02 Light chain, CH04 Heavy chain | | Authors: | Pancera, M, Louder, R, Mclellan, J.S, KWong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U2S
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain ZM109 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | McLellan, J.S, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U46
 
 | | CH04H/CH02L P212121 | | Descriptor: | CH02 Light chain Fab, CH04 Heavy chain Fab | | Authors: | Louder, R, Pancera, M, McLellan, J.S, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2011-12-21 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3QIC
 
 | | The structure of human glucokinase E339K mutation | | Descriptor: | GLYCEROL, Glucokinase, alpha-D-glucopyranose | | Authors: | Liu, Q, Liu, S, Liu, J. | | Deposit date: | 2011-01-27 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of E339K mutated human glucokinase reveals changes in the ATP binding site.
Febs Lett., 585, 2011
|
|
3PJZ
 
 | | Crystal Structure of the Potassium Transporter TrkH from Vibrio parahaemolyticus | | Descriptor: | POTASSIUM ION, Potassium uptake protein TrkH | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2010-11-10 | | Release date: | 2011-01-19 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | Crystal structure of a potassium ion transporter, TrkH.
Nature, 471, 2011
|
|
3QNQ
 
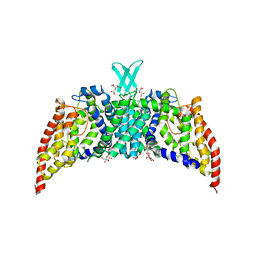 | | Crystal structure of the transporter ChbC, the IIC component from the N,N'-diacetylchitobiose-specific phosphotransferase system | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, PTS system, ... | | Authors: | Cao, Y, Jin, X, Huang, H, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2011-02-08 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Crystal structure of a phosphorylation-coupled saccharide transporter.
Nature, 473, 2011
|
|
6E58
 
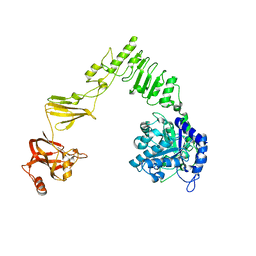 | | Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) | | Descriptor: | CALCIUM ION, Secreted Endo-beta-N-acetylglucosaminidase (EndoS) | | Authors: | Klontz, E.H, Trastoy, B, Gunther, S, Guerin, M.E, Sundberg, E.J. | | Deposit date: | 2018-07-19 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular Basis of Broad SpectrumN-Glycan Specificity and Processing of Therapeutic IgG Monoclonal Antibodies by Endoglycosidase S2.
ACS Cent Sci, 5, 2019
|
|
1B53
 
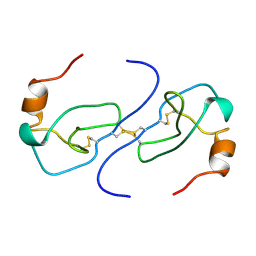 | | NMR STRUCTURE OF HUMAN MIP-1A D26A, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MIP-1A | | Authors: | Waltho, J.P, Higgins, L.D, Craven, C.J, Tan, P, Dudgeon, T. | | Deposit date: | 1999-01-11 | | Release date: | 1999-07-22 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES. Characterization of active disaggregated chemokine variants.
J.Biol.Chem., 274, 1999
|
|
1B50
 
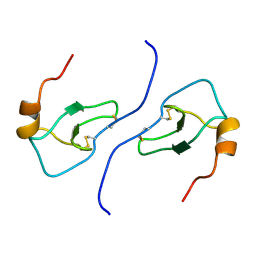 | | NMR STRUCTURE OF HUMAN MIP-1A D26A, 10 STRUCTURES | | Descriptor: | MIP-1A | | Authors: | Waltho, J.P, Higgins, L.D, Craven, C.J, Tan, P, Dudgeon, T. | | Deposit date: | 1999-01-11 | | Release date: | 1999-07-22 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1alpha, MIP-1beta, and RANTES. Characterization of active disaggregated chemokine variants.
J.Biol.Chem., 274, 1999
|
|
7Z5G
 
 | | human apo MATCAP | | Descriptor: | Uncharacterized protein KIAA0895-like | | Authors: | Bak, J, Adamoupolos, A, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-05-11 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
7Z6S
 
 | | MATCAP bound to a human 14 protofilament microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bak, J, Perrakis, A. | | Deposit date: | 2022-03-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
7Z5H
 
 | | human Zn MATCAP | | Descriptor: | Uncharacterized protein KIAA0895-like, ZINC ION | | Authors: | Bak, J, Adamopoulos, A, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
2M53
 
 | |
