1Y7Q
 
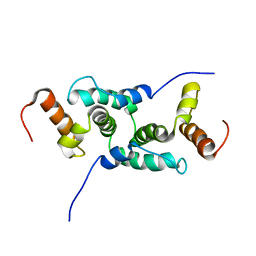 | | Mammalian SCAN domain dimer is a domain-swapped homologue of the HIV capsid C-terminal domain | | Descriptor: | Zinc finger protein 174 | | Authors: | Ivanov, D, Stone, J.R, Maki, J.L, Collins, T, Wagner, G. | | Deposit date: | 2004-12-09 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mammalian SCAN Domain Dimer Is a Domain-Swapped Homolog of the HIV Capsid C-Terminal Domain
Mol.Cell, 17, 2005
|
|
1IBX
 
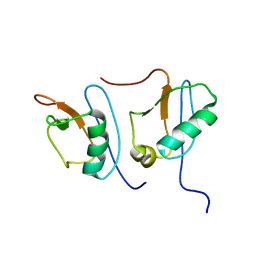 | | NMR STRUCTURE OF DFF40 AND DFF45 N-TERMINAL DOMAIN COMPLEX | | Descriptor: | CHIMERA OF IGG BINDING PROTEIN G AND DNA FRAGMENTATION FACTOR 45, DNA FRAGMENTATION FACTOR 40 | | Authors: | Zhou, P, Lugovskoy, A.A, McCarty, J.S, Li, P, Wagner, G. | | Deposit date: | 2001-03-29 | | Release date: | 2001-05-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of DFF40 and DFF45 N-terminal domain complex and mutual chaperone activity of DFF40 and DFF45.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1J8C
 
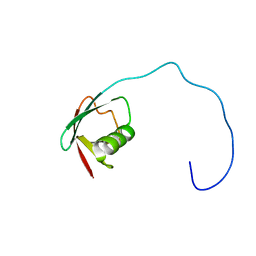 | | Solution Structure of the Ubiquitin-like Domain of hPLIC-2 | | Descriptor: | ubiquitin-like protein hPLIC-2 | | Authors: | Walters, K.J, Kleijnen, M.F, Goh, A.M, Wagner, G, Howley, P.M. | | Deposit date: | 2001-05-21 | | Release date: | 2002-03-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a.
Biochemistry, 41, 2002
|
|
1ZC1
 
 | | Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites | | Descriptor: | Ubiquitin fusion degradation protein 1 | | Authors: | Park, S, Isaacson, R, Kim, H.T, Silver, P.A, Wagner, G. | | Deposit date: | 2005-04-10 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Ufd1 Exhibits the AAA-ATPase Fold with Two Distinct Ubiquitin Interaction Sites
Structure, 13, 2005
|
|
2B86
 
 | |
1JJS
 
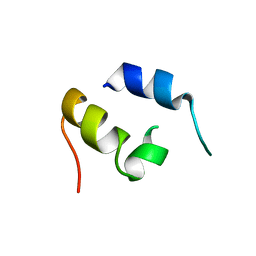 | | NMR Structure of IBiD, A Domain of CBP/p300 | | Descriptor: | CREB-BINDING PROTEIN | | Authors: | Lin, C.H, Hare, B.J, Wagner, G, Harrison, S.C, Maniatis, T, Fraenkel, E. | | Deposit date: | 2001-07-09 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A small domain of CBP/p300 binds diverse proteins: solution structure and functional studies.
Mol.Cell, 8, 2001
|
|
1JQ4
 
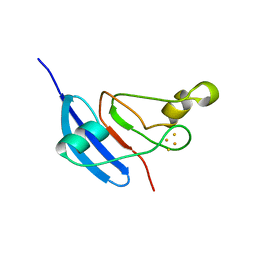 | | [2Fe-2S] Domain of Methane Monooxygenase Reductase from Methylococcus capsulatus (Bath) | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, METHANE MONOOXYGENASE COMPONENT C | | Authors: | Mueller, J, Lugovskoy, A.A, Wagner, G, Lippard, S.J. | | Deposit date: | 2001-08-03 | | Release date: | 2002-01-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the [2Fe-2S] ferredoxin domain from soluble methane monooxygenase reductase and interaction with its hydroxylase.
Biochemistry, 41, 2002
|
|
2AIV
 
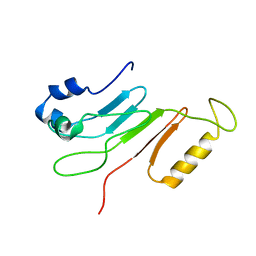 | | Multiple conformations in the ligand-binding site of the yeast nuclear pore targeting domain of NUP116P | | Descriptor: | fragment of Nucleoporin NUP116/NSP116 | | Authors: | Robinson, M.A, Park, S, Sun, Z.-Y.J, Silver, P, Wagner, G, Hogle, J. | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Multiple Conformations in the Ligand-binding Site of the Yeast Nuclear Pore-targeting Domain of Nup116p
J.Biol.Chem., 280, 2005
|
|
2BID
 
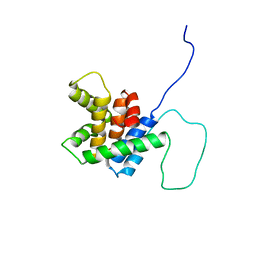 | | HUMAN PRO-APOPTOTIC PROTEIN BID | | Descriptor: | PROTEIN (BID) | | Authors: | Chou, J.J, Li, H, Salvesen, G.S, Yuan, J, Wagner, G. | | Deposit date: | 1999-01-27 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BID, an intracellular amplifier of apoptotic signaling.
Cell(Cambridge,Mass.), 96, 1999
|
|
1L3E
 
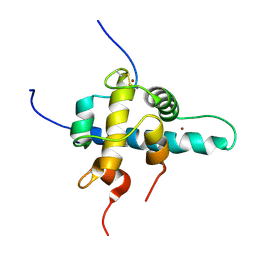 | | NMR Structures of the HIF-1alpha CTAD/p300 CH1 Complex | | Descriptor: | ZINC ION, hypoxia inducible factor-1 alpha subunit, p300 protein | | Authors: | Freedman, S.J, Sun, Z.J, Poy, F, Kung, A.L, Livingston, D.M, Wagner, G, Eck, M.J. | | Deposit date: | 2002-02-26 | | Release date: | 2002-04-24 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1L2Z
 
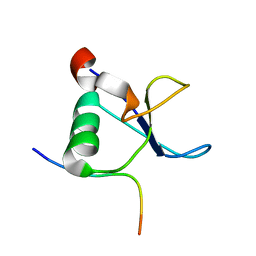 | | CD2BP2-GYF domain in complex with proline-rich CD2 tail segment peptide | | Descriptor: | CD2 ANTIGEN (CYTOPLASMIC TAIL)-BINDING PROTEIN 2, T-CELL SURFACE ANTIGEN CD2 | | Authors: | Freund, C, Kuhne, R, Yang, H, Park, S, Reinherz, E.L, Wagner, G. | | Deposit date: | 2002-02-26 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Dynamic interaction of CD2 with the GYF and the SH3 domain of compartmentalized effector molecules
Embo J., 21, 2002
|
|
1NFA
 
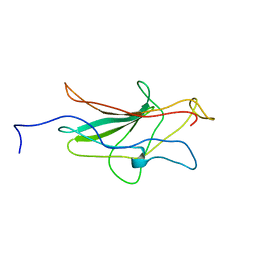 | | HUMAN TRANSCRIPTION FACTOR NFATC DNA BINDING DOMAIN, NMR, 10 STRUCTURES | | Descriptor: | HUMAN TRANSCRIPTION FACTOR NFATC1 | | Authors: | Wolfe, S.A, Zhou, P, Dotsch, V, Chen, L, You, A, Ho, S.N, Crabtree, G.R, Wagner, G, Verdine, G.L. | | Deposit date: | 1997-01-18 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Unusual Rel-like architecture in the DNA-binding domain of the transcription factor NFATc.
Nature, 385, 1997
|
|
5GCN
 
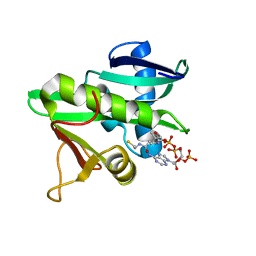 | | CATALYTIC DOMAIN OF TETRAHYMENA GCN5 HISTONE ACETYLTRANSFERASE IN COMPLEX WITH COENZYME A | | Descriptor: | COENZYME A, HISTONE ACETYLTRANSFERASE GCN5 | | Authors: | Lin, Y, Fletcher, C.M, Zhou, J, Allis, C.D, Wagner, G. | | Deposit date: | 1999-03-24 | | Release date: | 1999-07-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of GCN5 histone acetyltransferase bound to coenzyme A
Nature, 400, 1999
|
|
1N4Y
 
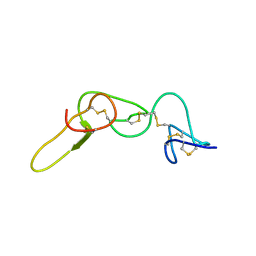 | | REFINED STRUCTURE OF KISTRIN | | Descriptor: | KISTRIN | | Authors: | Krezel, A.M, Krane, J, Dennis, M.S, Lazarus, R.A, Wagner, G. | | Deposit date: | 2002-11-02 | | Release date: | 2003-01-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kistrin, a potent platelet aggregation inhibitor and GP IIb-IIIa antagonist.
Science, 253, 1991
|
|
1KQR
 
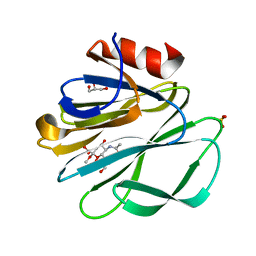 | | Crystal Structure of the Rhesus Rotavirus VP4 Sialic Acid Binding Domain in Complex with 2-O-methyl-alpha-D-N-acetyl neuraminic acid | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Dormitzer, P.R, Sun, Z.-Y.J, Wagner, G, Harrison, S.C. | | Deposit date: | 2002-01-07 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Rhesus Rotavirus VP4 Sialic Acid Binding Domain has a Galectin Fold with a
Novel Carbohydrate Binding Site
Embo J., 21, 2002
|
|
1NUI
 
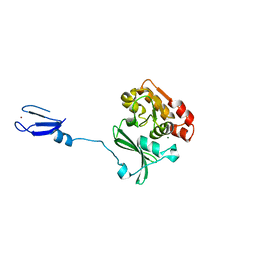 | | Crystal Structure of the primase fragment of Bacteriophage T7 primase-helicase protein | | Descriptor: | DNA primase/helicase, MAGNESIUM ION, ZINC ION | | Authors: | Kato, M, Ito, T, Wagner, G, Richardson, C.C, Ellenberger, T. | | Deposit date: | 2003-01-31 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Modular Architecture of the Bacteriophage T7 Primase Couples RNA primer Synthesis to DNA Synthesis
Mol.Cell, 11, 2003
|
|
4TQC
 
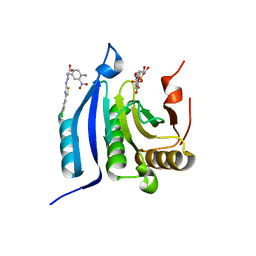 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2S)-3-(4-amino-3-nitrophenyl)-2-{2-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]hydrazinyl}propanoic acid, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic translation initiation factor 4E | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TQB
 
 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2E)-2-{2-[4-(4-bromophenyl)-1,3-thiazol-2-yl]hydrazinylidene}-3-(2-nitrophenyl)propanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4TPW
 
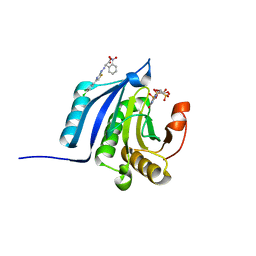 | | The co-complex structure of the translation initiation factor eIF4E with the inhibitor 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G | | Descriptor: | (2E)-2-{2-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]hydrazinylidene}-3-(2-nitrophenyl)propanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papadopoulos, E, Jenni, S, Wagner, G. | | Deposit date: | 2014-06-09 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2JOG
 
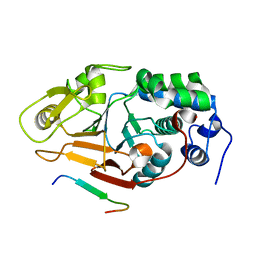 | | Structure of the calcineurin-NFAT complex | | Descriptor: | Calmodulin-dependent calcineurin A subunit alpha isoform, NFAT | | Authors: | Takeuchi, K, Roehrl, M.H, Sun, Z.Y, Wagner, G. | | Deposit date: | 2007-03-10 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the Calcineurin-NFAT Complex: Defining a T Cell Activation Switch Using Solution NMR and Crystal Coordinates.
Structure, 15, 2007
|
|
2IF1
 
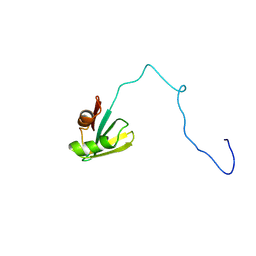 | | HUMAN TRANSLATION INITIATION FACTOR EIF1, NMR, 29 STRUCTURES | | Descriptor: | EIF1 | | Authors: | Fletcher, C.M, Hellen, C.U.T, Pestova, T.V, Wagner, G. | | Deposit date: | 1998-08-04 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and interactions of the translation initiation factor eIF1.
EMBO J., 18, 1999
|
|
2K4T
 
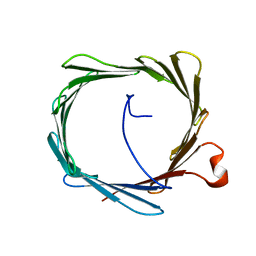 | | Solution structure of human VDAC-1 in LDAO micelles | | Descriptor: | Voltage-dependent anion-selective channel protein 1 | | Authors: | Hiller, S, Garces, R.G, Malia, T.J, Orekhov, V.Y, Colombini, M, Wagner, G. | | Deposit date: | 2008-06-17 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the integral human membrane protein VDAC-1 in detergent micelles.
Science, 321, 2008
|
|
2KD2
 
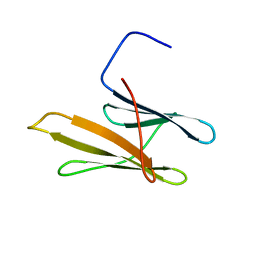 | | NMR Structure of FAIM-CTD | | Descriptor: | Fas apoptotic inhibitory molecule 1 | | Authors: | Hemond, M, Wagner, G. | | Deposit date: | 2009-01-01 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Fas apoptosis inhibitory molecule contains a novel beta-sandwich in contact with a partially ordered domain.
J.Mol.Biol., 386, 2009
|
|
2KY6
 
 | | Structure of ARC92VBD/MED25ACID | | Descriptor: | Mediator of RNA polymerase II transcription subunit 25 | | Authors: | Milbradt, A.G, Sun, Z.J, Selenko, P, Takeuchi, K, Naar, A.M, Wagner, G. | | Deposit date: | 2010-05-14 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the VP16 transactivator target in the Mediator.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2GUT
 
 | | Solution structure of the trans-activation domain of the human co-activator ARC105 | | Descriptor: | ARC/MEDIATOR, Positive cofactor 2 glutamine/Q-rich-associated protein | | Authors: | Vought, B.W, Jim Sun, Z.-Y, Hyberts, S.G, Wagner, G, Naar, A.M. | | Deposit date: | 2006-05-01 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis.
Nature, 442, 2006
|
|
