2AL6
 
 | | FERM domain of Focal Adhesion Kinase | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Ceccarelli, D.F, Song, H.K, Poy, F, Schaller, M.D, Eck, M.J. | | Deposit date: | 2005-08-04 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the FERM Domain of Focal Adhesion Kinase
J.Biol.Chem., 281, 2006
|
|
2DS7
 
 | | Structure of the ZBD in the hexagonal crystal form | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
6L18
 
 | |
6LHN
 
 | | RLGSGG-AtPRT6 UBR box | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Kwon, D.H, Song, H.K. | | Deposit date: | 2019-12-09 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Use of the LC3B-fusion technique for biochemical and structural studies of proteins involved in the N-degron pathway.
J.Biol.Chem., 295, 2020
|
|
1QZY
 
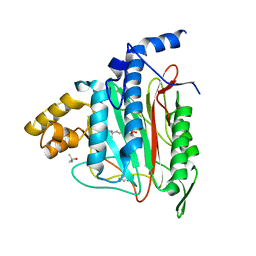 | | Human Methionine Aminopeptidase in complex with bengamide inhibitor LAF153 and cobalt | | Descriptor: | (E)-(2R,3R,4S,5R)-3,4,5-TRIHYDROXY-2-METHOXY-8,8-DIMETHYL-NON-6-ENOIC ACID ((3S,6R)-6-HYDROXY-2-OXO-AZEPAN-3-YL)-AMIDE, COBALT (II) ION, Methionine aminopeptidase 2, ... | | Authors: | Eck, M.J, Song, H.K, Morollo, A. | | Deposit date: | 2003-09-18 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proteomics-based target identification: bengamides as a new class of methionine aminopeptidase inhibitors.
J.Biol.Chem., 278, 2003
|
|
1M27
 
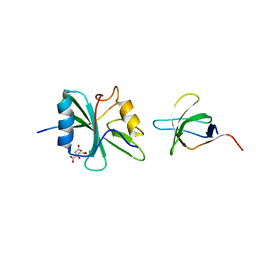 | | Crystal structure of SAP/FynSH3/SLAM ternary complex | | Descriptor: | CITRATE ANION, Proto-oncogene tyrosine-protein kinase FYN, SH2 domain protein 1A, ... | | Authors: | Chan, B, Griesbach, J, Song, H.K, Poy, F, Terhorst, C, Eck, M.J. | | Deposit date: | 2002-06-21 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | SAP couples Fyn to SLAM immune receptors.
NAT.CELL BIOL., 5, 2003
|
|
1OIL
 
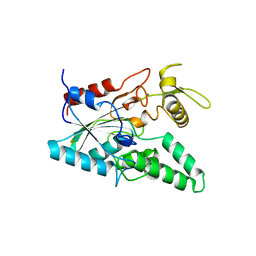 | | STRUCTURE OF LIPASE | | Descriptor: | CALCIUM ION, LIPASE | | Authors: | Kim, K.K, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 1996-12-06 | | Release date: | 1997-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor.
Structure, 5, 1997
|
|
1NTV
 
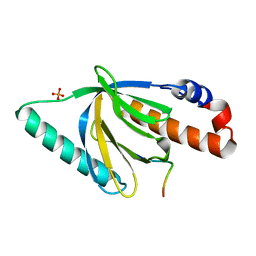 | | Crystal Structure of the Disabled-1 (Dab1) PTB domain-ApoER2 peptide complex | | Descriptor: | Apolipoprotein E Receptor-2 peptide, Disabled homolog 1, PHOSPHATE ION | | Authors: | Stolt, P.C, Jeon, H, Song, H.K, Herz, J, Eck, M.J, Blacklow, S.C. | | Deposit date: | 2003-01-30 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Origins of Peptide Selectivity and Phosphoinositide Binding Revealed by Structures of Disabled-1 PTB Domain Complexes
Structure, 11, 2003
|
|
1NHL
 
 | | SNAP-23N Structure | | Descriptor: | Synaptosomal-associated protein 23 | | Authors: | Freedman, S.J, Song, H.K, Xu, Y, Eck, M.J. | | Deposit date: | 2002-12-19 | | Release date: | 2003-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Homotetrameric Structure of the SNAP-23 N-terminal Coiled-coil Domain
J.Biol.Chem., 278, 2003
|
|
1NU2
 
 | | Crystal structure of the murine Disabled-1 (Dab1) PTB domain-ApoER2 peptide-PI-4,5P2 ternary complex | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Disabled homolog 1, peptide derived from murine Apolipoprotein E Receptor-2 | | Authors: | Stolt, P.C, Jeon, H, Song, H.K, Herz, J, Eck, M.J, Blacklow, S.C. | | Deposit date: | 2003-01-30 | | Release date: | 2003-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Origins of Peptide Selectivity and Phosphoinositide Binding Revealed by Structures of Disabled-1 PTB Domain Complexes
Structure, 11, 2003
|
|
4FBA
 
 | | Structure of mutant RIP from barley seeds in complex with adenine | | Descriptor: | ADENINE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FBB
 
 | | Structure of mutant RIP from barley seeds in complex with adenine (AMP-incubated) | | Descriptor: | ADENINE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3PO0
 
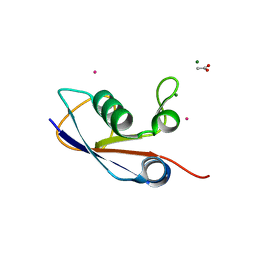 | | Crystal structure of SAMP1 from Haloferax volcanii | | Descriptor: | ACETATE ION, CADMIUM ION, MAGNESIUM ION, ... | | Authors: | Jeong, Y.J, Jeong, B.-C, Song, H.K. | | Deposit date: | 2010-11-21 | | Release date: | 2011-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of ubiquitin-like small archaeal modifier protein 1 (SAMP1) from Haloferax volcanii.
Biochem.Biophys.Res.Commun., 405, 2011
|
|
4FBH
 
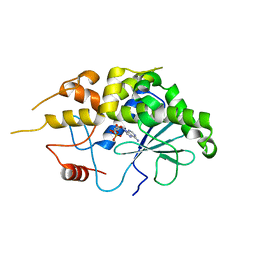 | | Structure of RIP from barley seeds | | Descriptor: | ADENOSINE MONOPHOSPHATE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FB9
 
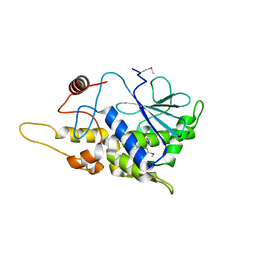 | | Structure of mutant RIP from barley seeds | | Descriptor: | Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4GQW
 
 | |
4HAN
 
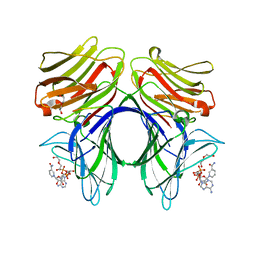 | | Crystal structure of Galectin 8 with NDP52 peptide | | Descriptor: | Calcium-binding and coiled-coil domain-containing protein 2, DI(HYDROXYETHYL)ETHER, Galectin-8, ... | | Authors: | Kim, B.-W, Hong, S.B, Kim, J.H, Kwon, D.H, Song, H.K. | | Deposit date: | 2012-09-27 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural basis for recognition of autophagic receptor NDP52 by the sugar receptor galectin-8.
Nat Commun, 4, 2013
|
|
4FBC
 
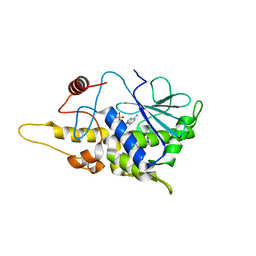 | | Structure of mutant RIP from barley seeds in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Protein synthesis inhibitor I | | Authors: | Lee, B.-G, Kim, M.K, Suh, S.W, Song, H.K. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the ribosome-inactivating protein from barley seeds reveal a unique activation mechanism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4GQV
 
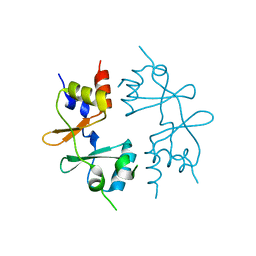 | | Crystal structure of CBS-pair protein, CBSX1 from Arabidopsis thaliana | | Descriptor: | CBS domain-containing protein CBSX1, chloroplastic | | Authors: | Jeong, B.-C, Park, S.H, Yoo, K.S, Shin, J.S, Song, H.K. | | Deposit date: | 2012-08-24 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Crystal structure of the single cystathionine beta-synthase domain-containing protein CBSX1 from Arabidopsis thaliana
Biochem.Biophys.Res.Commun., 430, 2013
|
|
4GQY
 
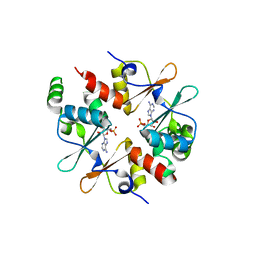 | | Crystal structure of CBSX2 in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CBS domain-containing protein CBSX2, chloroplastic | | Authors: | Jeong, B.C, Song, H.K. | | Deposit date: | 2012-08-24 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Change in single cystathionine beta-synthase domain-containing protein from a bent to flat conformation upon adenosine monophosphate binding
J.Struct.Biol., 183, 2013
|
|
4EBR
 
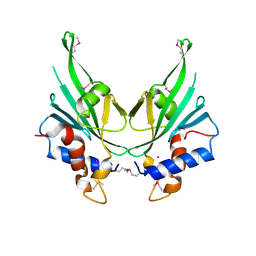 | | Crystal structure of Autophagic E2, Atg10 | | Descriptor: | MERCURY (II) ION, Ubiquitin-like-conjugating enzyme ATG10 | | Authors: | Hong, S.B, Kim, B.W, Kim, J.H, Song, H.K. | | Deposit date: | 2012-03-24 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Structure of the autophagic E2 enzyme Atg10
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3RUJ
 
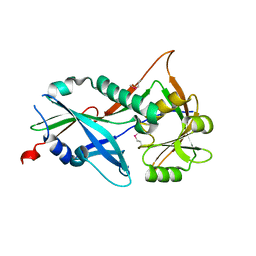 | |
3RUI
 
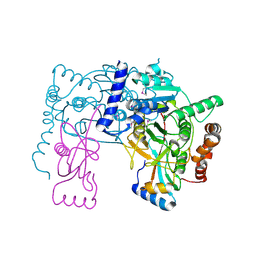 | | Crystal structure of Atg7C-Atg8 complex | | Descriptor: | Autophagy-related protein 8, Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Hong, S.B, Kim, B.W, Song, H.K. | | Deposit date: | 2011-05-05 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4HNZ
 
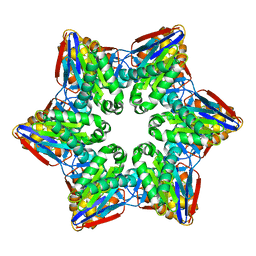 | |
4HO7
 
 | |
